Model Insights and Interactive Analysis Introduction
Wallaroo provides the ability to perform interactive analysis so organizations can explore the data from a pipeline and learn how the data is behaving. With this information and the knowledge of your particular business use case you can then choose appropriate thresholds for persistent automatic assays as desired.
IMPORTANT NOTE
Model insights operates over time and is difficult to demo in a notebook without pre-canned data. We assume you have an active pipeline that has been running and making predictions over time and show you the code you may use to analyze your pipeline.
Monitoring tasks called assays monitors a model’s predictions or the data coming into the model against an established baseline. Changes in the distribution of this data can be an indication of model drift, or of a change in the environment that the model trained for. This can provide tips on whether a model needs to be retrained or the environment data analyzed for accuracy or other needs.
Assay Details
Assays contain the following attributes:
| Attribute | Default | Description |
|---|---|---|
| Name | The name of the assay. Assay names must be unique. | |
| Baseline Data | Data that is known to be “typical” (typically distributed) and can be used to determine whether the distribution of new data has changed. | |
| Schedule | Every 24 hours at 1 AM | Configure the start time and frequency of when the new analysis will run. New assays are configured to run a new analysis for every 24 hours starting at the end of the baseline period. This period can be configured through the SDK. |
| Group Results | Daily | How the results are grouped: Daily (Default), Every Minute, Weekly, or Monthly. |
| Metric | PSI | Population Stability Index (PSI) is an entropy-based measure of the difference between distributions. Maximum Difference of Bins measures the maximum difference between the baseline and current distributions (as estimated using the bins). Sum of the difference of bins sums up the difference of occurrences in each bin between the baseline and current distributions. |
| Threshold | 0.1 | Threshold for deciding the difference between distributions is similar(small) or different(large), as evaluated by the metric. The default of 0.1 is generally a good threshold when using PSI as the metric. |
| Number of Bins | 5 | Number of bins used to partition the baseline data. By default, the binning scheme is percentile (quantile) based. The binning scheme can be configured (see Bin Mode, below). Note that the total number of bins will include the set number plus the left_outlier and the right_outlier, so the total number of bins will be the total set + 2. |
| Bin Mode | Quantile | Specify the Binning Scheme. Available options are: Quantile binning defines the bins using percentile ranges (each bin holds the same percentage of the baseline data). Equal binning defines the bins using equally spaced data value ranges, like a histogram. Custom allows users to set the range of values for each bin, with the Left Outlier always starting at Min (below the minimum values detected from the baseline) and the Right Outlier always ending at Max (above the maximum values detected from the baseline). |
| Bin Weight | Equally Weighted | The weight applied to each bin. The bin weights can be either set to Equally Weighted (the default) where each bin is weighted equally, or Custom where the bin weights can be adjusted depending on which are considered more important for detecting model drift. |
Manage Assays via the Wallaroo Dashboard
Assays can be created and used via the Wallaroo Dashboard.
Accessing Assays Through the Pipeline Dashboard
Assays created through the Wallaroo Dashboard are accessed through the Pipeline Dashboard through the following process.
- Log into the Wallaroo Dashboard.
- Select the workspace containing the pipeline with the models being monitored from the Change Current Workspace and Workspace Management drop down.
- Select View Pipelines.
- Select the pipeline containing the models being monitored.
- Select Insights.
The Wallaroo Assay Dashboard contains the following elements. For more details of each configuration type, see the Model Insights and Assays Introduction.
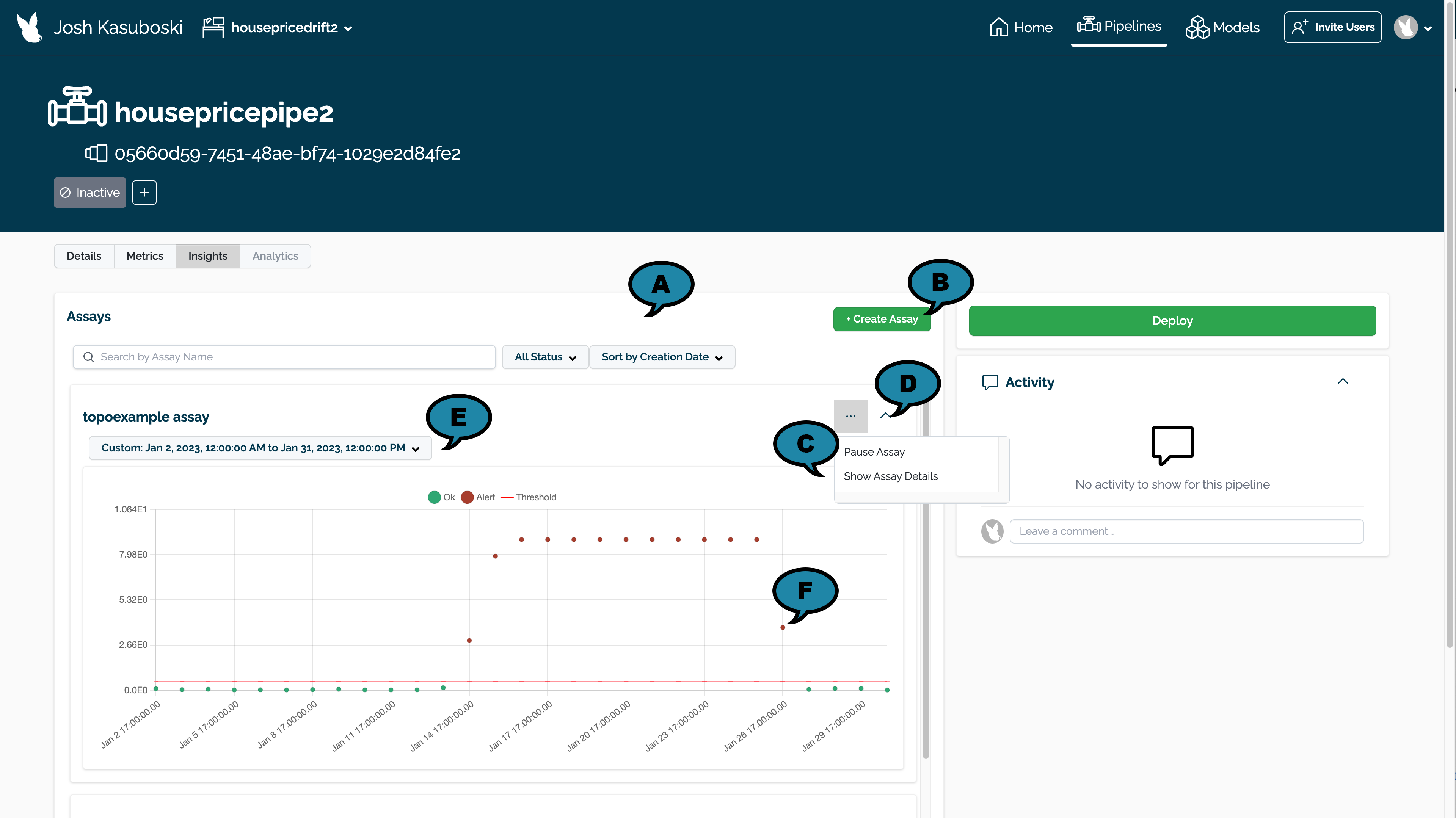
- (A) Filter Assays: Filter assays by the following:
- Name
- Status:
- Active: The assay is currently running.
- Paused: The assay is paused until restarted.
- Drift Detected: One or more drifts have been detected.
- Sort By
- Sort by Creation Date: Sort by the most recent Assays first.
- Last Assay Run: Sort by the most recent Assay Last Run date.
- (B) Create Assay: Create a new assay.
- (C) Assay Controls:
- Pause/Start Assay: Pause a running assay, or start one that was paused.
- Show Assay Details: View assay details. See Assay Details View for more details.
- (D) Collapse Assay: Collapse or Expand the assay for view.
- (E) Time Period for Assay Data: Set the time period for data to be used in displaying the assay results.
- (F) Assay Events: Select an individual assay event to see more details. See View Assay Alert Details for more information.
Assay Details View
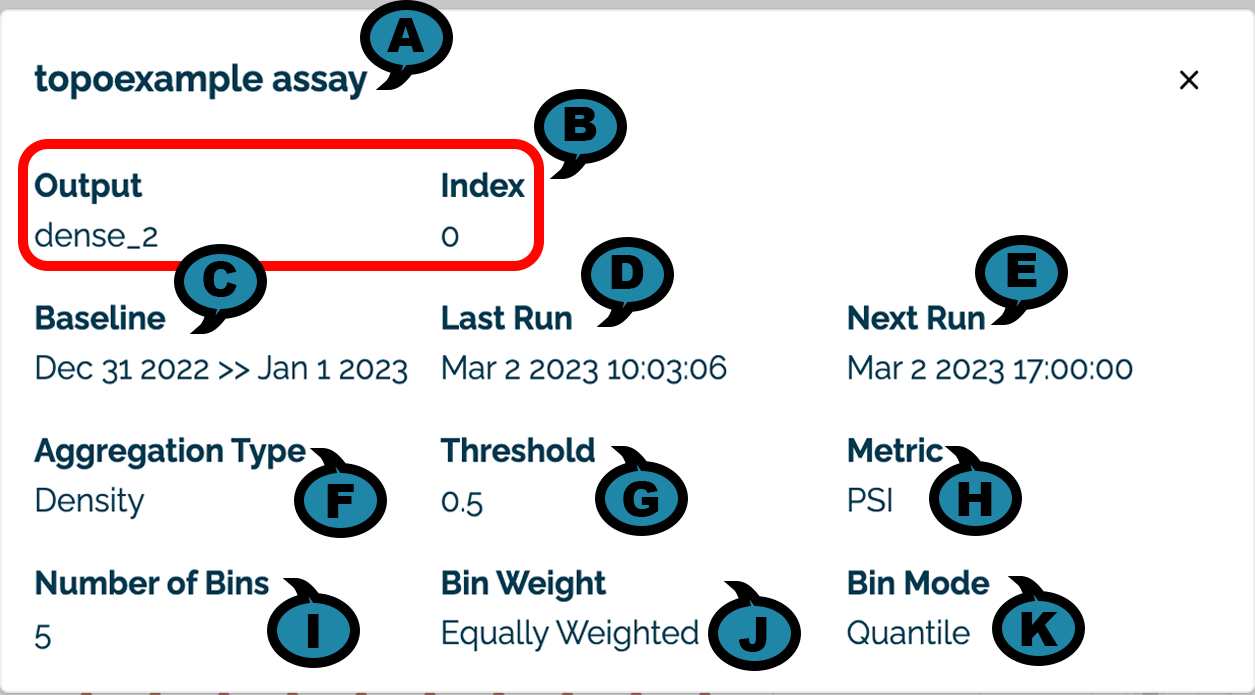
The following details are visible by selecting the Assay View Details icon:
- (A) Assay Name: The name of the assay displayed.
- (B) Input / Output: The input or output and the index of the element being monitored.
- (C) Baseline: The time period used to generate the baseline. For baselines generated from a file, the baseline displayed
Uploaded File. - (D) Last Run: The date and time the assay was last run.
- (E) Next Run: The future date and time the assay will be run again. NOTE: If the assay is paused, then it will not run at the scheduled time. When unpaused, the date will be updated to the next date and time that the assay will be run.
- (F) Aggregation Type: The aggregation type used with the assay.
- (G) Threshold: The threshold value used for the assay.
- (H) Metric: The metric type used for the assay.
- (I) Number of Bins: The number of bins used for the assay.
- (J) Bin Weight: The weight applied to each bin.
- (K) Bin Mode: The type of bin node applied to each bin.
View Assay Alert Details
To view details on an assay alert:
- Select the data with available alert data.
- Mouse hover of a specific Assay Event Alert to view the data and time of the event and the alert value.
- Select the Assay Event Alert to view the Baseline and Window details of the alert including the left_outlier and right_outlier.
Hover over a bar chart graph to view additional details.
- Select the ⊗ symbol to exit the Assay Event Alert details and return to the Assay View.
Build an Assay Through the Pipeline Dashboard
To create a new assay through the Wallaroo Pipeline Dashboard:
- Log into the Wallaroo Dashboard.
- Select the workspace containing the pipeline with the models being monitored from the Change Current Workspace and Workspace Management drop down.
- Select View Pipelines.
- Select the pipeline containing the models being monitored.
- Select Insights.
- Select +Create Assay.
- On the Create Assay module, enter the following:
On the Assay Name section, enter the following:
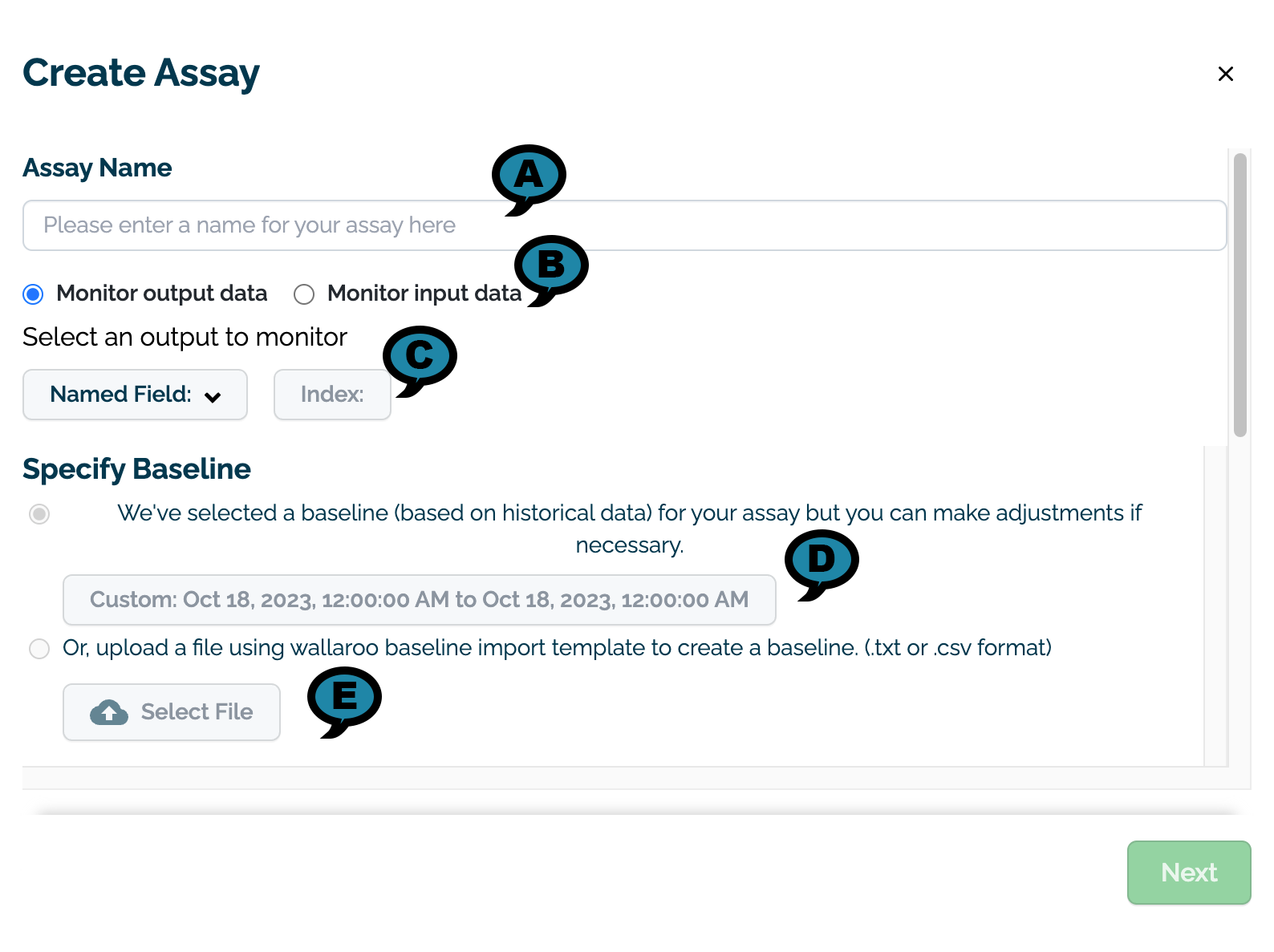
Assay Name (A): The name of the new assay.
Monitor output data or Monitor input data (B): Select whether to monitor input or output data.
Select an output/input to monitor (C): Select the input or output to monitor.
- Named Field: The name of the field to monitor.
- Index: The index of the monitored field.
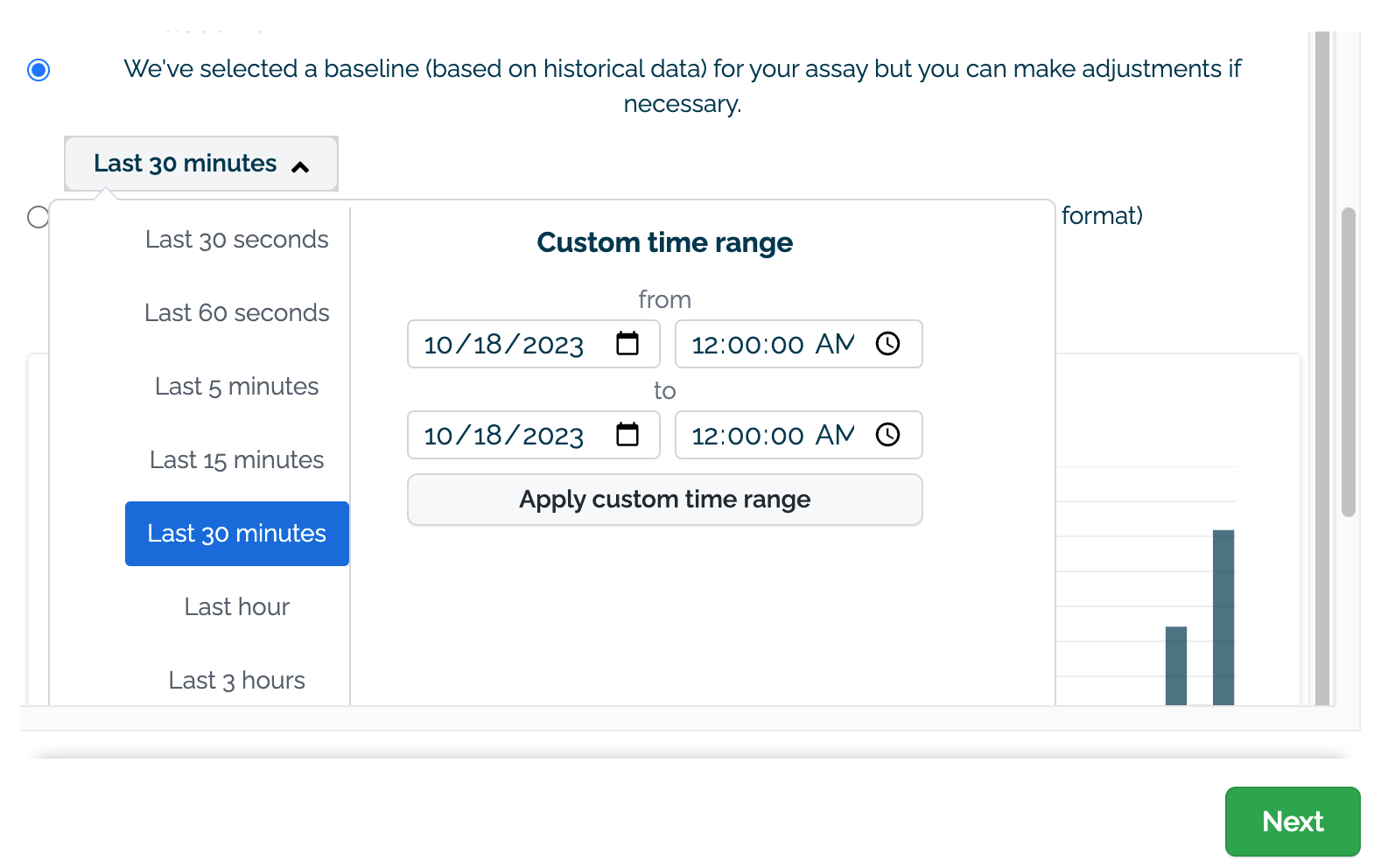
On the Specify Baseline section, select one of the following options:
- (D) Select the data to use for the baseline. This can either be set with a preset recent time period (last 30 seconds, last 60 seconds, etc) or with a custom date range.
(E) Upload an assay baseline file as either a CSV or TXT file. These assay baselines must be a list of numpy (aka float) values that are comma and newline separated, terminating at the last record with no additional commas or returns.
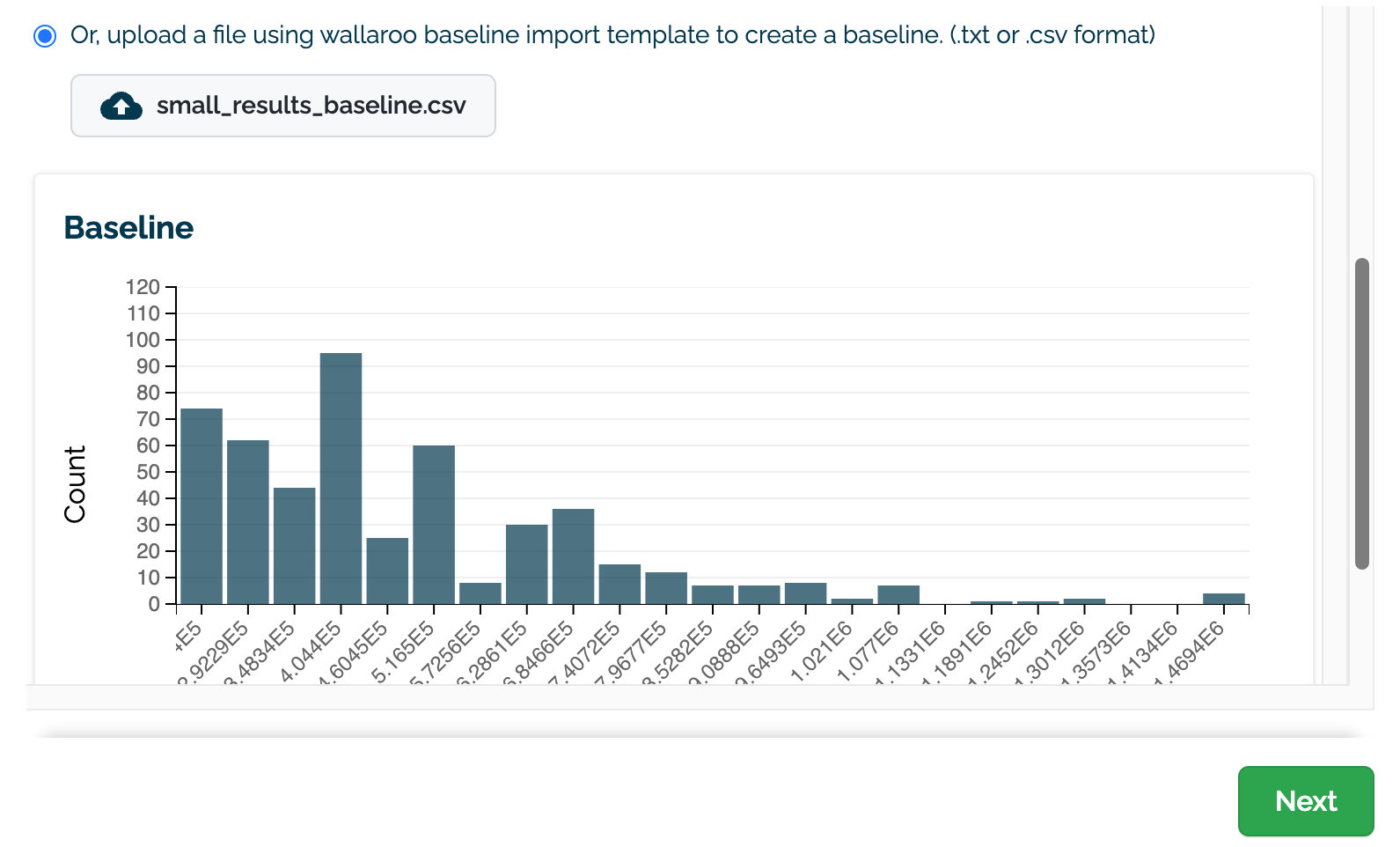
For example:
684577.200, 921561.500, 705013.440, 725875.900, 684577.200, 379398.300, 266405.600, 256630.310
Once selected, a preview graph of the baseline values will be displayed (C). Note that this may take a few seconds to generate.
Select Next to continue.
- On the Settings Module:
Set the date and time range to view values generated by the assay. This can either be set with a preset recent time period (last 30 seconds, last 60 seconds, etc) or with a custom date range.
New assays are configured to run a new analysis for every 24 hours starting at the end of the baseline period. For information on how to adjust the scheduling period and other settings for the assay scheduling window, see the SDK section on how to Schedule Assay.
Set the following Advanced Settings.
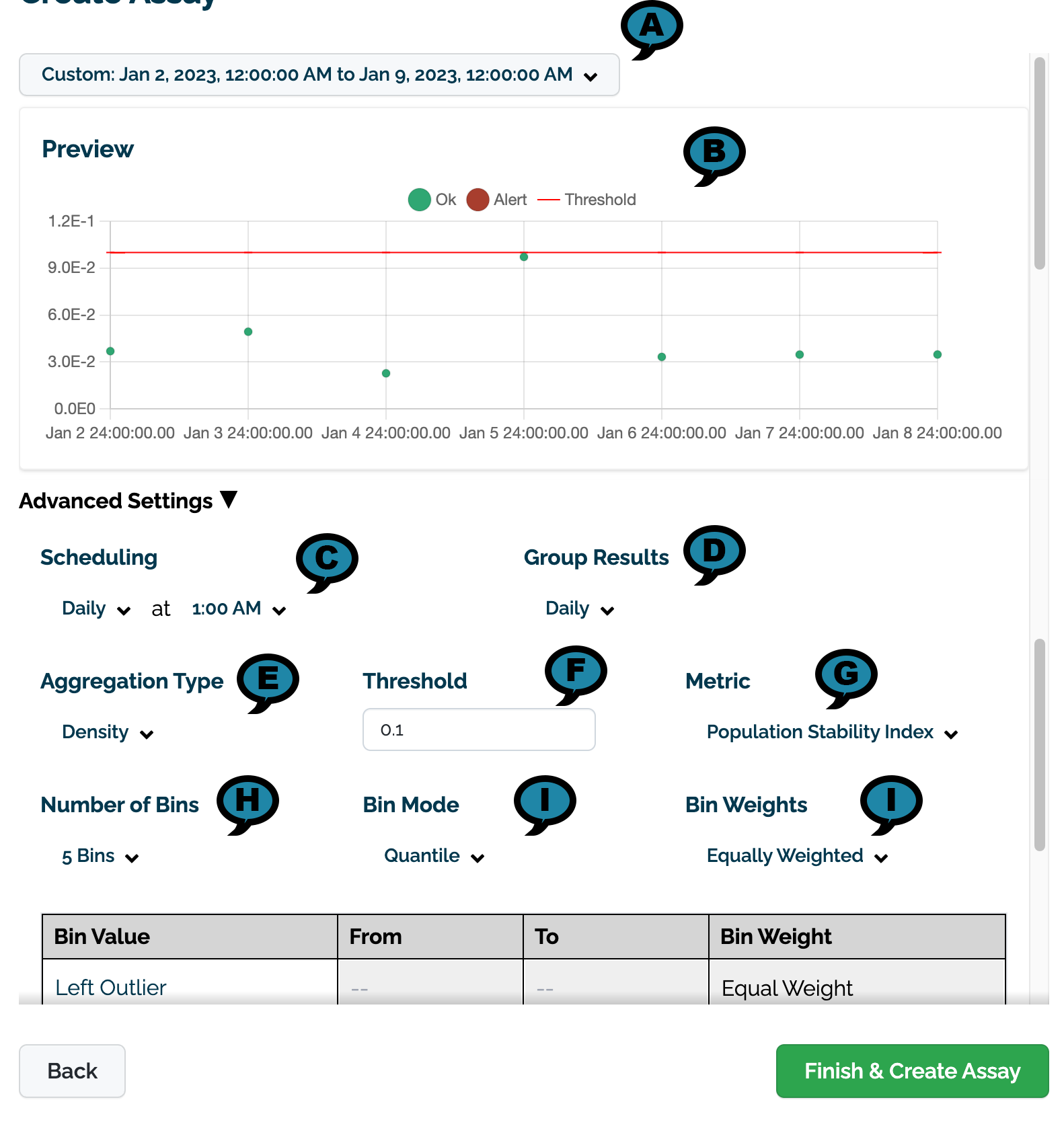
- (A) Preview Date Range: The date and times to for the preview chart.
- (B) Preview: A preview of the assay results will be displayed based on the settings below.
- (C) Scheduling: Set the Frequency (Daily, Every Minute, Hourly, Weekly, Default: Daily) and the Time (increments of one hour Default: 1:00 AM).
- (D) Group Results: How the results are grouped: Daily (Default), Every Minute, Weekly, or Monthly.
- (E) Aggregation Type: Density or Cumulative.
- (F) Threshold:
- Default: 0.1
- (G) Metric:
- Default: Population Stability Index
- Maximum Difference of Bins
- Sum of the Difference of Bins
- (H) Number of Bins: From 5 to 14. Default: 5
- (F) Bin Mode:
- Equally Spaced
- Default: Quantile
- (I) Bin Weights: The bin weights:
- Equally Weighted (Default)
- Custom: Users can assign their own bin weights as required.
Review the preview chart to verify the settings are correct.
Select Build to complete the process and build the new assay.
Once created, it may take a few minutes for the assay to complete compiling data. If needed, reload the Pipeline Dashboard to view changes.
Model Insights via the Wallaroo Dashboard SDK
Assays generated through the Wallaroo SDK can be previewed, configured, and uploaded to the Wallaroo Ops instance. The following is a condensed version of this process. For full details see the Wallaroo SDK Essentials Guide: Assays Management guide.
Model drift detection with assays using the Wallaroo SDK follows this general process.
- Define the Baseline: From either historical inference data for a specific model in a pipeline, or from a pre-determine array of data, a baseline is formed.
- Assay Preview: Once the baseline is formed, we preview the assay and configure the different options until we have the the best method of detecting environment or model drift.
- Create Assay: With the previews and configuration complete, we upload the assay. The assay will perform an analysis on a regular scheduled based on the configuration.
- Get Assay Results: Retrieve the analyses and use them to detect model drift and possible sources.
- Pause/Resume Assay: Pause or restart an assay as needed.
Define the Baseline
Assay baselines are defined with the wallaroo.client.build_assay method. Through this process we define the baseline from either a range of dates or pre-generated values.
wallaroo.client.build_assay take the following parameters:
| Parameter | Type | Description |
|---|---|---|
| assay_name | String (Required) - required | The name of the assay. Assay names must be unique across the Wallaroo instance. |
| pipeline | wallaroo.pipeline.Pipeline (Required) | The pipeline the assay is monitoring. |
| model_name | String (Required) | The name of the model to monitor. |
| iopath | String (Required) | The input/output data for the model being tracked in the format input/output field index. Only one value is tracked for any assay. For example, to track the output of the model’s field house_value at index 0, the iopath is 'output house_value 0. |
| baseline_start | datetime.datetime (Optional) | The start time for the inferences to use as the baseline. Must be included with baseline_end. Cannot be included with baseline_data. |
| baseline_end | datetime.datetime (Optional) | The end time of the baseline window. the baseline. Windows start immediately after the baseline window and are run at regular intervals continuously until the assay is deactivated or deleted. Must be included with baseline_start. Cannot be included with baseline_data.. |
| baseline_data | numpy.array (Optional) | The baseline data in numpy array format. Cannot be included with either baseline_start or baseline_data. |
Baselines are created in one of two ways:
- Date Range: The
baseline_startandbaseline_endretrieves the inference requests and results for the pipeline from the start and end period. This data is summarized and used to create the baseline. - Numpy Values: The
baseline_datasets the baseline from a provided numpy array.
Define the Baseline Example
This example shows two methods of defining the baseline for an assay:
"assays from date baseline": This assay uses historical inference requests to define the baseline. This assay is saved to the variableassay_builder_from_dates."assays from numpy": This assay uses a pre-generated numpy array to define the baseline. This assay is saved to the variableassay_builder_from_numpy.
In both cases, the following parameters are used:
| Parameter | Value |
|---|---|
| assay_name | "assays from date baseline" and "assays from numpy" |
| pipeline | mainpipeline: A pipeline with a ML model that predicts house prices. The output field for this model is variable. |
| model_name | "houseprice-predictor" - the model name set during model upload. |
| iopath | These assays monitor the model’s output field variable at index 0. From this, the iopath setting is "output variable 0". |
The difference between the two assays’ parameters determines how the baseline is generated.
"assays from date baseline": Uses thebaseline_startandbaseline_endto set the time period of inference requests and results to gather data from."assays from numpy": Uses a pre-generated numpy array as for the baseline data.
For each of our assays, we will set the time period of inference data to compare against the baseline data.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
# assay builder by baseline
assay_builder_from_numpy = wl.build_assay(assay_name="assays from numpy",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_data = small_results_baseline)
# set the width, interval, and time period
assay_builder_from_numpy.add_run_until(datetime.datetime.now())
assay_builder_from_numpy.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_numpy = assay_builder_from_numpy.build()
assay_results_from_numpy = assay_config_from_numpy.interactive_run()
Baseline DataFrame
The method wallaroo.assay_config.AssayBuilder.baseline_dataframe returns a DataFrame of the assay baseline generated from the provided parameters. This includes:
metadata: The inference metadata with the model information, inference time, and other related factors.indata: Each input field assigned with the labelin.{input field name}.outdata: Each output field assigned with the labelout.{output field name}
Note that for assays generated from numpy values, there is only the out data based on the supplied baseline data.
In the following example, the baseline DataFrame is retrieved.
display(assay_builder_from_dates.baseline_dataframe())
| time | metadata | input_tensor_0 | input_tensor_1 | input_tensor_2 | input_tensor_3 | input_tensor_4 | input_tensor_5 | input_tensor_6 | input_tensor_7 | ... | input_tensor_9 | input_tensor_10 | input_tensor_11 | input_tensor_12 | input_tensor_13 | input_tensor_14 | input_tensor_15 | input_tensor_16 | input_tensor_17 | output_variable_0 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1708013922866 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [49610, 341497], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 4.0 | 3.00 | 3710.0 | 20000.0 | 2.0 | 0.0 | 2.0 | 5.0 | ... | 2760.0 | 950.0 | 47.669600 | -122.261000 | 3970.0 | 20000.0 | 79.0 | 0.0 | 0.0 | 1.514079e+06 |
| 1 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 3.0 | 2.50 | 1500.0 | 7420.0 | 1.0 | 0.0 | 0.0 | 3.0 | ... | 1000.0 | 500.0 | 47.723598 | -122.174004 | 1840.0 | 7272.0 | 42.0 | 0.0 | 0.0 | 4.196772e+05 |
| 2 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 4.0 | 2.50 | 2009.0 | 5000.0 | 2.0 | 0.0 | 0.0 | 3.0 | ... | 2009.0 | 0.0 | 47.257702 | -122.197998 | 2009.0 | 5182.0 | 0.0 | 0.0 | 0.0 | 3.208637e+05 |
| 3 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 3.0 | 1.75 | 1530.0 | 7245.0 | 1.0 | 0.0 | 0.0 | 4.0 | ... | 1530.0 | 0.0 | 47.730999 | -122.191002 | 1530.0 | 7490.0 | 31.0 | 0.0 | 0.0 | 4.319292e+05 |
| 4 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 3.0 | 1.75 | 1480.0 | 4800.0 | 2.0 | 0.0 | 0.0 | 4.0 | ... | 1140.0 | 340.0 | 47.656700 | -122.397003 | 1810.0 | 4800.0 | 70.0 | 0.0 | 0.0 | 5.361757e+05 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 496 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 4.0 | 2.25 | 2560.0 | 12100.0 | 1.0 | 0.0 | 0.0 | 4.0 | ... | 1760.0 | 800.0 | 47.631001 | -122.108002 | 2240.0 | 12100.0 | 38.0 | 0.0 | 0.0 | 7.019407e+05 |
| 497 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 2.0 | 1.00 | 1160.0 | 5000.0 | 1.0 | 0.0 | 0.0 | 4.0 | ... | 1160.0 | 0.0 | 47.686501 | -122.399002 | 1750.0 | 5000.0 | 77.0 | 0.0 | 0.0 | 4.508677e+05 |
| 498 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 4.0 | 2.50 | 1910.0 | 5000.0 | 2.0 | 0.0 | 0.0 | 3.0 | ... | 1910.0 | 0.0 | 47.360802 | -122.036003 | 2020.0 | 5000.0 | 9.0 | 0.0 | 0.0 | 2.962027e+05 |
| 499 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 3.0 | 1.50 | 1590.0 | 8911.0 | 1.0 | 0.0 | 0.0 | 3.0 | ... | 1590.0 | 0.0 | 47.739399 | -122.251999 | 1590.0 | 9625.0 | 58.0 | 0.0 | 0.0 | 4.371780e+05 |
| 500 | 1708013983808 | {'last_model': '{"model_name":"house-price-estimator","model_sha":"e22a0831aafd9917f3cc87a15ed267797f80e2afa12ad7d8810ca58f173b8cc6"}', 'pipeline_version': '', 'elapsed': [2517407, 2287534], 'dropped': [], 'partition': 'engine-58b565bf45-29xnm'} | 4.0 | 2.75 | 2640.0 | 4000.0 | 2.0 | 0.0 | 0.0 | 5.0 | ... | 1730.0 | 910.0 | 47.672699 | -122.296997 | 1530.0 | 3740.0 | 89.0 | 0.0 | 0.0 | 7.184457e+05 |
501 rows × 21 columns
assay_builder_from_numpy.baseline_dataframe()
| output_variable_0 | |
|---|---|
| 0 | 419677.20 |
| 1 | 320863.72 |
| 2 | 431929.20 |
| 3 | 536175.70 |
| 4 | 343304.63 |
| ... | ... |
| 495 | 701940.70 |
| 496 | 450867.70 |
| 497 | 296202.70 |
| 498 | 437177.97 |
| 499 | 718445.70 |
500 rows × 1 columns
Baseline Stats
The method wallaroo.assay.AssayAnalysis.baseline_stats() returns a pandas.core.frame.DataFrame of the baseline stats.
The baseline stats for each assay are displayed in the examples below.
assay_results_from_dates[0].baseline_stats()
| Baseline | |
|---|---|
| count | 501 |
| min | 236238.671875 |
| max | 1514079.375 |
| mean | 495193.231786 |
| median | 442168.125 |
| std | 226075.814267 |
| start | None |
| end | None |
assay_results_from_numpy[0].baseline_stats()
| Baseline | |
|---|---|
| count | 500 |
| min | 236238.67 |
| max | 1489624.3 |
| mean | 493155.46054 |
| median | 441840.425 |
| std | 221657.583536 |
| start | None |
| end | None |
Baseline Bins
The method wallaroo.assay.AssayAnalysis.baseline_bins a simple dataframe to with the edge/bin data for a baseline.
assay_results_from_dates[0].baseline_bins()
| b_edges | b_edge_names | b_aggregated_values | b_aggregation | |
|---|---|---|---|---|
| 0 | 2.362387e+05 | left_outlier | 0.000000 | Density |
| 1 | 2.962027e+05 | q_20 | 0.203593 | Density |
| 2 | 4.159643e+05 | q_40 | 0.195609 | Density |
| 3 | 4.640602e+05 | q_60 | 0.203593 | Density |
| 4 | 6.821819e+05 | q_80 | 0.197605 | Density |
| 5 | 1.514079e+06 | q_100 | 0.199601 | Density |
| 6 | inf | right_outlier | 0.000000 | Density |
assay_results_from_numpy[0].baseline_bins()
| b_edges | b_edge_names | b_aggregated_values | b_aggregation | |
|---|---|---|---|---|
| 0 | 236238.67 | left_outlier | 0.000 | Density |
| 1 | 296202.70 | q_20 | 0.204 | Density |
| 2 | 415964.30 | q_40 | 0.196 | Density |
| 3 | 464057.38 | q_60 | 0.200 | Density |
| 4 | 675545.44 | q_80 | 0.200 | Density |
| 5 | 1489624.30 | q_100 | 0.200 | Density |
| 6 | inf | right_outlier | 0.000 | Density |
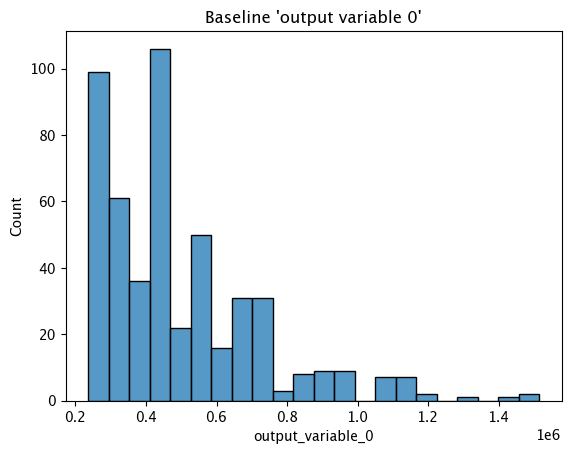
Baseline Histogram Chart
The method wallaroo.assay_config.AssayBuilder.baseline_histogram returns a histogram chart of the assay baseline generated from the provided parameters.
assay_builder_from_dates.baseline_histogram()
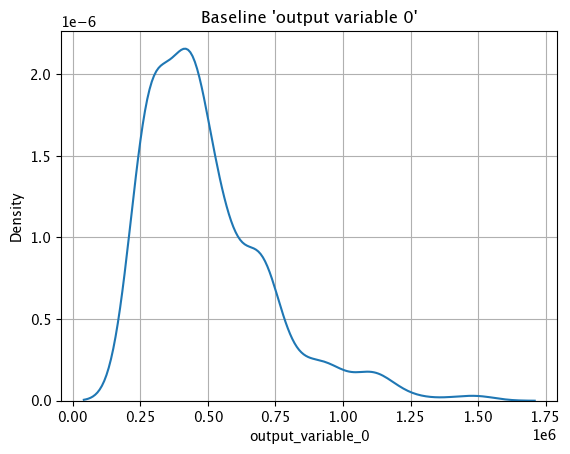
Baseline KDE Chart
The method wallaroo.assay_config.AssayBuilder.baseline_kde returns a Kernel Density Estimation (KDE) chart of the assay baseline generated from the provided parameters.
assay_builder_from_dates.baseline_kde()
Baseline ECDF Chart
The method wallaroo.assay_config.AssayBuilder.baseline_ecdf returns a Empirical Cumulative Distribution Function (CDF) chart of the assay baseline generated from the provided parameters.
assay_builder_from_dates.baseline_ecdf()
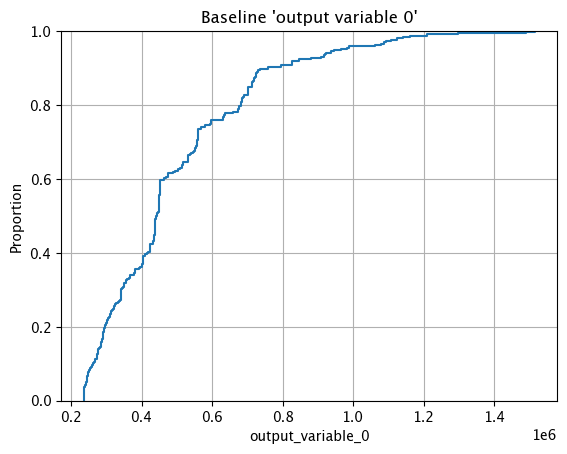
Assay Preview
Now that the baseline is defined, we look at different configuration options and view how the assay baseline and results changes. Once we determine what gives us the best method of determining model drift, we can create the assay.
Analysis List Chart Scores
Analysis List scores show the assay scores for each assay result interval in one chart. Values that are outside of the alert threshold are colored red, while scores within the alert threshold are green.
Assay chart scores are displayed with the method wallaroo.assay.AssayAnalysisList.chart_scores(title: Optional[str] = None), with ability to display an optional title with the chart.
The following example shows retrieving the assay results and displaying the chart scores. From our example, we have two windows - the first should be green, and the second is red showing that values were outside the alert threshold.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.chart_scores()

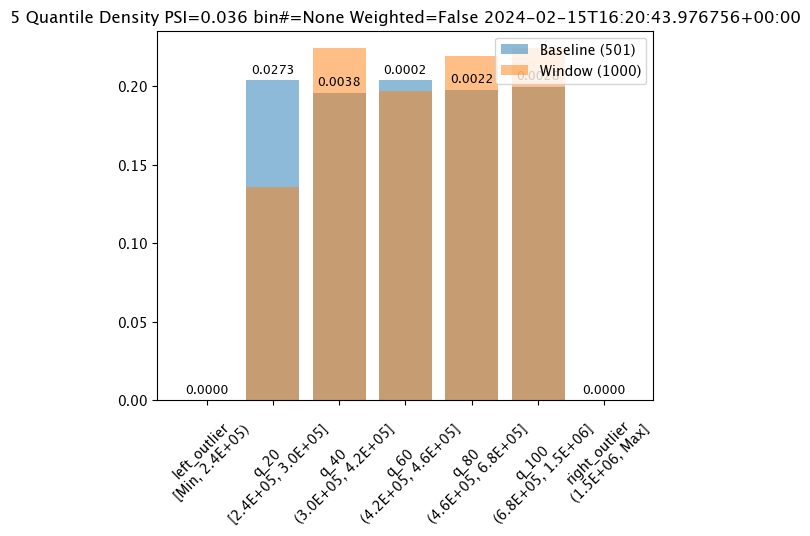
Analysis Chart
The method wallaroo.assay.AssayAnalysis.chart() displays a comparison between the baseline and an interval of inference data.
This is compared to the Chart Scores, which is a list of all of the inference data split into intervals, while the Analysis Chart shows the breakdown of one set of inference data against the baseline.
Score from the Analysis List Chart Scores and each element from the Analysis List DataFrame generates
The following fields are included.
| Field | Type | Description |
|---|---|---|
| baseline mean | Float | The mean of the baseline values. |
| window mean | Float | The mean of the window values. |
| baseline median | Float | The median of the baseline values. |
| window median | Float | The median of the window values. |
| bin_mode | String | The binning mode used for the assay. |
| aggregation | String | The aggregation mode used for the assay. |
| metric | String | The metric mode used for the assay. |
| weighted | Bool | Whether the bins were manually weighted. |
| score | Float | The score from the assay window. |
| scores | List(Float) | The score from each assay window bin. |
| index | Integer/None | The window index. Interactive assay runs are None. |
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
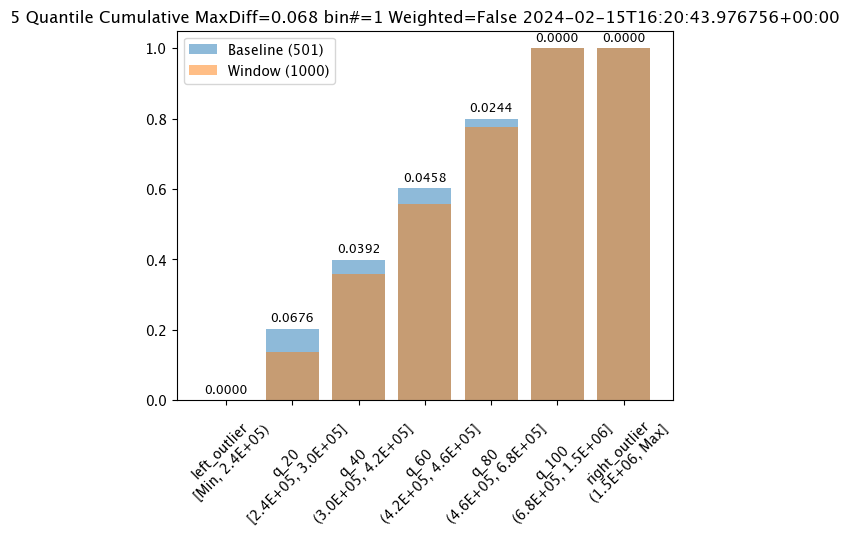
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0363497101644573
scores = [0.0, 0.027271477163285655, 0.003847844548034077, 0.000217023993714693, 0.002199485350158766, 0.0028138791092641195, 0.0]
index = None
Analysis List DataFrame
wallaroo.assay.AssayAnalysisList.to_dataframe() returns a DataFrame showing the assay results for each window aka individual analysis. This DataFrame contains the following fields:
| Field | Type | Description |
|---|---|---|
| assay_id | Integer/None | The assay id. Only provided from uploaded and executed assays. |
| name | String/None | The name of the assay. Only provided from uploaded and executed assays. |
| iopath | String/None | The iopath of the assay. Only provided from uploaded and executed assays. |
| score | Float | The assay score. |
| start | DateTime | The DateTime start of the assay window. |
| min | Float | The minimum value in the assay window. |
| max | Float | The maximum value in the assay window. |
| mean | Float | The mean value in the assay window. |
| median | Float | The median value in the assay window. |
| std | Float | The standard deviation value in the assay window. |
| warning_threshold | Float/None | The warning threshold of the assay window. |
| alert_threshold | Float/None | The alert threshold of the assay window. |
| status | String | The assay window status. Values are:
|
For this example, the assay analysis list DataFrame is listed.
From this tutorial, we should have 2 windows of dta to look at, each one minute apart. The first window should show status: OK, with the second window with the very large house prices will show status: alert
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.to_dataframe()
| assay_id | name | iopath | score | start | min | max | mean | median | std | warning_threshold | alert_threshold | status | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | 0.036350 | 2024-02-15T16:20:43.976756+00:00 | 2.362387e+05 | 1489624.250 | 5.177634e+05 | 4.486278e+05 | 227729.030050 | None | 0.25 | Ok | ||
| 1 | None | 8.868614 | 2024-02-15T16:22:43.976756+00:00 | 1.514079e+06 | 2016006.125 | 1.885772e+06 | 1.946438e+06 | 160046.727324 | None | 0.25 | Alert |
Analysis List Full DataFrame
wallaroo.assay.AssayAnalysisList.to_full_dataframe() returns a DataFrame showing all values, including the inputs and outputs from the assay results for each window aka individual analysis. This DataFrame contains the following fields:
pipeline_id warning_threshold bin_index created_at
| Field | Type | Description |
|---|---|---|
| window_start | DateTime | The date and time when the window period began. |
| analyzed_at | DateTime | The date and time when the assay analysis was performed. |
| elapsed_millis | Integer | How long the analysis took to perform in milliseconds. |
| baseline_summary_count | Integer | The number of data elements from the baseline. |
| baseline_summary_min | Float | The minimum value from the baseline summary. |
| baseline_summary_max | Float | The maximum value from the baseline summary. |
| baseline_summary_mean | Float | The mean value of the baseline summary. |
| baseline_summary_median | Float | The median value of the baseline summary. |
| baseline_summary_std | Float | The standard deviation value of the baseline summary. |
| baseline_summary_edges_{0…n} | Float | The baseline summary edges for each baseline edge from 0 to number of edges. |
| summarizer_type | String | The type of summarizer used for the baseline. See wallaroo.assay_config for other summarizer types. |
| summarizer_bin_weights | List / None | If baseline bin weights were provided, the list of those weights. Otherwise, None. |
| summarizer_provided_edges | List / None | If baseline bin edges were provided, the list of those edges. Otherwise, None. |
| status | String | The assay window status. Values are:
|
| id | Integer/None | The id for the window aka analysis. Only provided from uploaded and executed assays. |
| assay_id | Integer/None | The assay id. Only provided from uploaded and executed assays. |
| pipeline_id | Integer/None | The pipeline id. Only provided from uploaded and executed assays. |
| warning_threshold | Float | The warning threshold set for the assay. |
| warning_threshold | Float | The warning threshold set for the assay. |
| bin_index | Integer/None | The bin index for the window aka analysis. |
| created_at | Datetime/None | The date and time the window aka analysis was generated. Only provided from uploaded and executed assays. |
For this example, full DataFrame from an assay preview is generated.
From this tutorial, we should have 2 windows of dta to look at, each one minute apart. The first window should show status: OK, with the second window with the very large house prices will show status: alert
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.to_full_dataframe()
| window_start | analyzed_at | elapsed_millis | baseline_summary_count | baseline_summary_min | baseline_summary_max | baseline_summary_mean | baseline_summary_median | baseline_summary_std | baseline_summary_edges_0 | ... | summarizer_type | summarizer_bin_weights | summarizer_provided_edges | status | id | assay_id | pipeline_id | warning_threshold | bin_index | created_at | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2024-02-15T16:20:43.976756+00:00 | 2024-02-15T16:26:42.266029+00:00 | 82 | 501 | 236238.671875 | 1514079.375 | 495193.231786 | 442168.125 | 226075.814267 | 236238.671875 | ... | UnivariateContinuous | None | None | Ok | None | None | None | None | None | None |
| 1 | 2024-02-15T16:22:43.976756+00:00 | 2024-02-15T16:26:42.266134+00:00 | 83 | 501 | 236238.671875 | 1514079.375 | 495193.231786 | 442168.125 | 226075.814267 | 236238.671875 | ... | UnivariateContinuous | None | None | Alert | None | None | None | None | None | None |
2 rows × 86 columns
Analysis Compare Basic Stats
The method wallaroo.assay.AssayAnalysis.compare_basic_stats returns a DataFrame comparing one set of inference data against the baseline.
This is compared to the Analysis List DataFrame, which is a list of all of the inference data split into intervals, while the Analysis Compare Basic Stats shows the breakdown of one set of inference data against the baseline.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].compare_basic_stats()
| Baseline | Window | diff | pct_diff | |
|---|---|---|---|---|
| count | 501.0 | 1000.0 | 499.000000 | 99.600798 |
| min | 236238.671875 | 236238.671875 | 0.000000 | 0.000000 |
| max | 1514079.375 | 1489624.25 | -24455.125000 | -1.615181 |
| mean | 495193.231786 | 517763.394625 | 22570.162839 | 4.557850 |
| median | 442168.125 | 448627.8125 | 6459.687500 | 1.460912 |
| std | 226075.814267 | 227729.03005 | 1653.215783 | 0.731266 |
| start | None | 2024-02-15T16:20:43.976756+00:00 | NaN | NaN |
| end | None | 2024-02-15T16:21:43.976756+00:00 | NaN | NaN |
Configure Assays
Before creating the assay, configure the assay and continue to preview it until the best method for detecting drift is set. The following options are available.
Score Metric
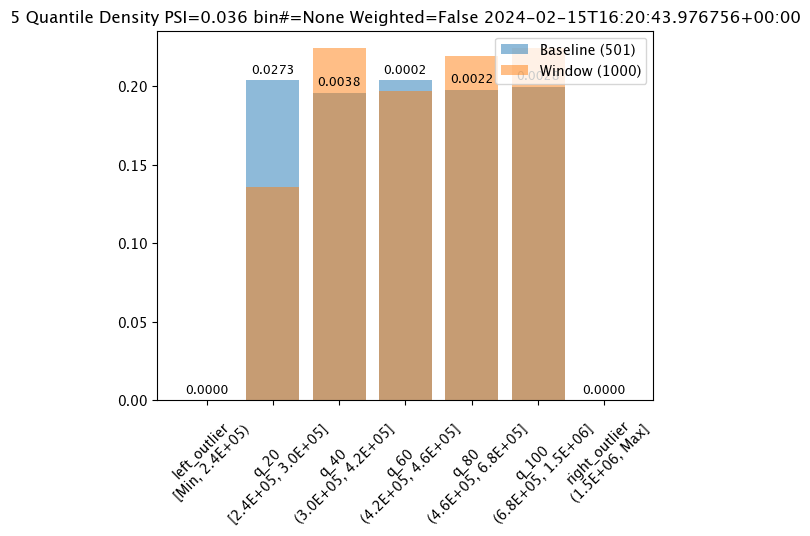
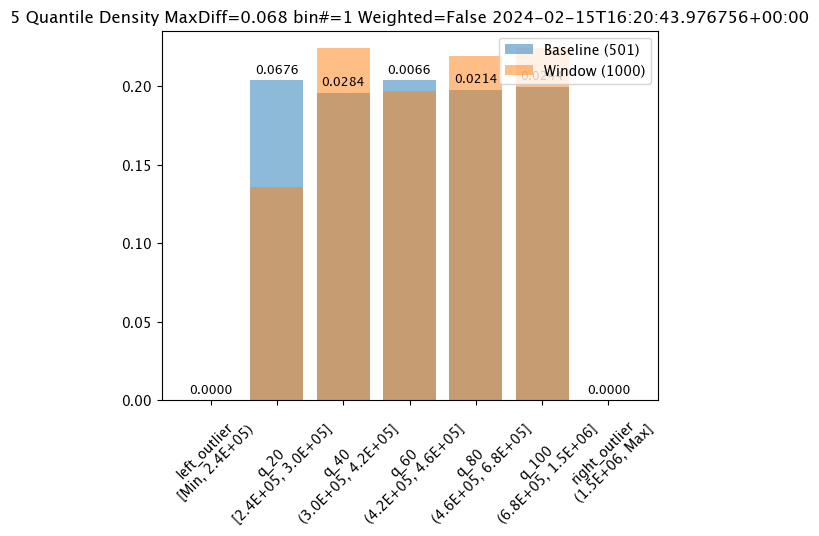
The score is a distance between the baseline and the analysis window. The larger the score, the greater the difference between the baseline and the analysis window. The following methods are provided determining the score:
PSI(Default) - Population Stability Index (PSI).MAXDIFF: Maximum difference between corresponding bins.SUMDIFF: Mum of differences between corresponding bins.
The metric type used is updated with the wallaroo.assay_config.AssayBuilder.add_metric(metric: wallaroo.assay_config.Metric) method.
The following three charts use each of the metrics. Note how the scores change based on the score type used.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set metric PSI mode
assay_builder_from_dates.summarizer_builder.add_metric(wallaroo.assay_config.Metric.PSI)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0363497101644573
scores = [0.0, 0.027271477163285655, 0.003847844548034077, 0.000217023993714693, 0.002199485350158766, 0.0028138791092641195, 0.0]
index = None
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set metric MAXDIFF mode
assay_builder_from_dates.summarizer_builder.add_metric(wallaroo.assay_config.Metric.MAXDIFF)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Density
metric = MaxDiff
weighted = False
score = 0.06759281437125747
scores = [0.0, 0.06759281437125747, 0.028391217564870255, 0.006592814371257472, 0.02139520958083832, 0.02439920159680639, 0.0]
index = 1
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set metric SUMDIFF mode
assay_builder_from_dates.summarizer_builder.add_metric(wallaroo.assay_config.Metric.SUMDIFF)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Density
metric = SumDiff
weighted = False
score = 0.07418562874251496
scores = [0.0, 0.06759281437125747, 0.028391217564870255, 0.006592814371257472, 0.02139520958083832, 0.02439920159680639, 0.0]
index = None
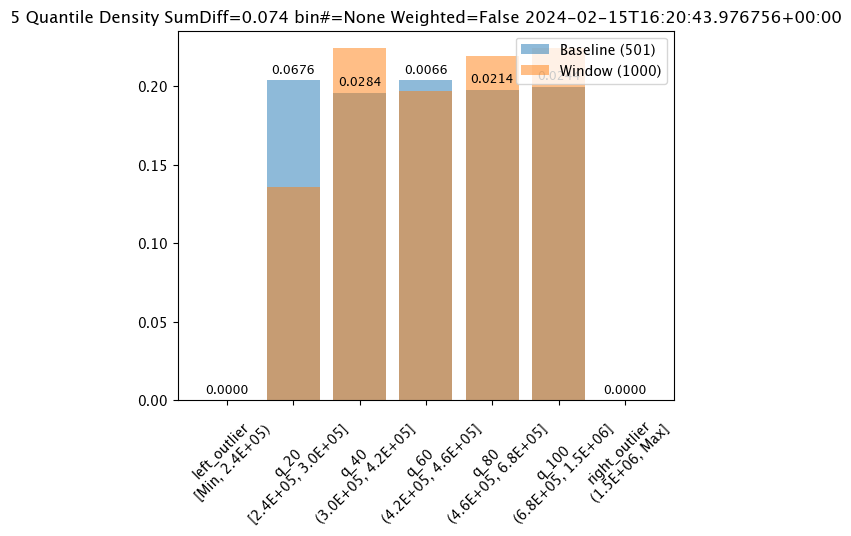
Alert Threshold
Assay alert thresholds are modified with the wallaroo.assay_config.AssayBuilder.add_alert_threshold(alert_threshold: float) method. By default alert thresholds are 0.1.
The following example updates the alert threshold to 0.5.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
assay_builder_from_dates.add_alert_threshold(0.5)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.to_dataframe()
| assay_id | name | iopath | score | start | min | max | mean | median | std | warning_threshold | alert_threshold | status | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | 0.036350 | 2024-02-15T16:20:43.976756+00:00 | 2.362387e+05 | 1489624.250 | 5.177634e+05 | 4.486278e+05 | 227729.030050 | None | 0.5 | Ok | ||
| 1 | None | 8.868614 | 2024-02-15T16:22:43.976756+00:00 | 1.514079e+06 | 2016006.125 | 1.885772e+06 | 1.946438e+06 | 160046.727324 | None | 0.5 | Alert |
Number of Bins
Number of bins sets how the baseline data is partitioned. The total number of bins includes the set number plus the left_outlier and the right_outlier, so the total number of bins will be the total set + 2.
The number of bins is set with the wallaroo.assay_config.UnivariateContinousSummarizerBuilder.add_num_bins(num_bins: int) method.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# Set the number of bins
# update number of bins here
assay_builder_from_dates.summarizer_builder.add_num_bins(10)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.05250979748389363
scores = [0.0, 0.009076998929542533, 0.01924002322223739, 0.0021945246367443406, 0.0016700458183385653, 0.005779503770625584, 0.002393429678215835, 0.002942858220315506, 0.00010651192741915124, 0.00046961759334670583, 0.008636283687108028, 0.0]
index = None
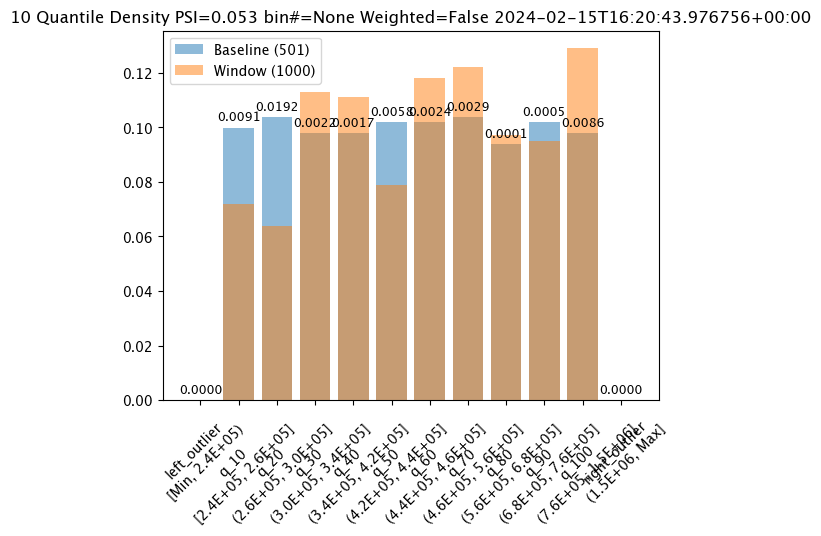
Binning Mode
Binning Mode defines how the bins are separated. Binning modes are modified through the wallaroo.assay_config.UnivariateContinousSummarizerBuilder.add_bin_mode(bin_mode: bin_mode: wallaroo.assay_config.BinMode, edges: Optional[List[float]] = None).
Available bin_mode values from wallaroo.assay_config.Binmode are the following:
QUANTILE(Default): Based on percentages. Ifnum_binsis 5 then quintiles so bins are created at the 20%, 40%, 60%, 80% and 100% points.EQUAL: Evenly spaced bins where each bin is set with the formulamin - max / num_binsPROVIDED: The user provides the edge points for the bins.
If PROVIDED is supplied, then a List of float values must be provided for the edges parameter that matches the number of bins.
The following examples are used to show how each of the binning modes effects the bins.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# update binning mode here
assay_builder_from_dates.summarizer_builder.add_bin_mode(wallaroo.assay_config.BinMode.QUANTILE)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0363497101644573
scores = [0.0, 0.027271477163285655, 0.003847844548034077, 0.000217023993714693, 0.002199485350158766, 0.0028138791092641195, 0.0]
index = None
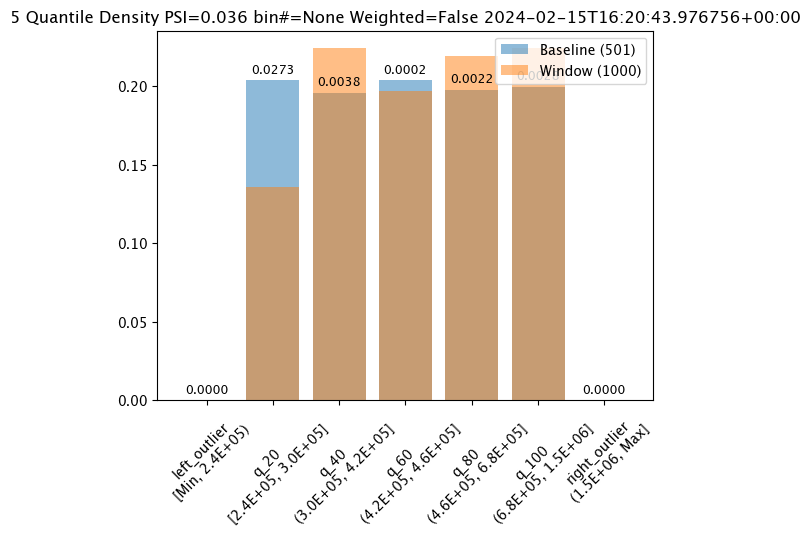
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# update binning mode here
assay_builder_from_dates.summarizer_builder.add_bin_mode(wallaroo.assay_config.BinMode.EQUAL)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Equal
aggregation = Density
metric = PSI
weighted = False
score = 0.013362603453760629
scores = [0.0, 0.0016737762070682225, 1.1166481947075492e-06, 0.011233704798893194, 1.276169365380064e-07, 0.00045387818266796784, 0.0]
index = None
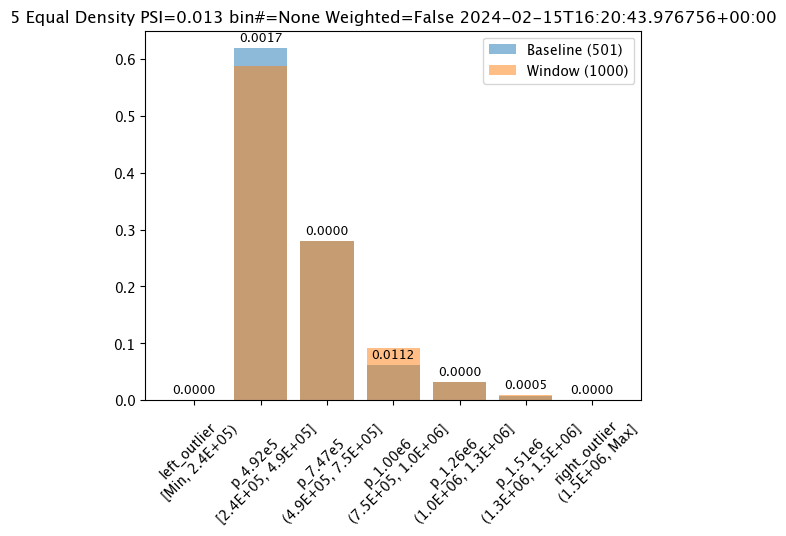
The following example manually sets the bin values.
The values in this dataset run from 200000 to 1500000. We can specify the bins with the BinMode.PROVIDED and specifying a list of floats with the right hand / upper edge of each bin and optionally the lower edge of the smallest bin. If the lowest edge is not specified the threshold for left outliers is taken from the smallest value in the baseline dataset.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
edges = [200000.0, 400000.0, 600000.0, 800000.0, 1500000.0, 2000000.0]
# update binning mode here
assay_builder_from_dates.summarizer_builder.add_bin_mode(wallaroo.assay_config.BinMode.PROVIDED, edges)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Provided
aggregation = Density
metric = PSI
weighted = False
score = 0.01005936099521711
scores = [0.0, 0.0030207963288415803, 0.00011480201840874194, 0.00045327555974347976, 0.0037119550613212583, 0.0027585320269020493, 0.0]
index = None
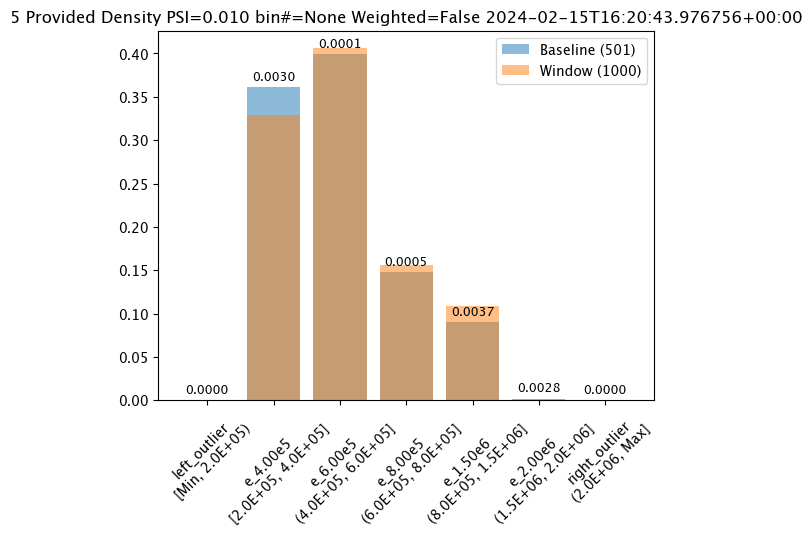
Aggregation Options
Assay aggregation options are modified with the wallaroo.assay_config.AssayBuilder.add_aggregation(aggregation: wallaroo.assay_config.Aggregation) method. The following options are provided:
Aggregation.DENSITY(Default): Count the number/percentage of values that fall in each bin.Aggregation.CUMULATIVE: Empirical Cumulative Density Function style, which keeps a cumulative count of the values/percentages that fall in each bin.
The following example demonstrate the different results between the two.
#Aggregation.DENSITY - the default
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
assay_builder_from_dates.summarizer_builder.add_aggregation(wallaroo.assay_config.Aggregation.DENSITY)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0363497101644573
scores = [0.0, 0.027271477163285655, 0.003847844548034077, 0.000217023993714693, 0.002199485350158766, 0.0028138791092641195, 0.0]
index = None
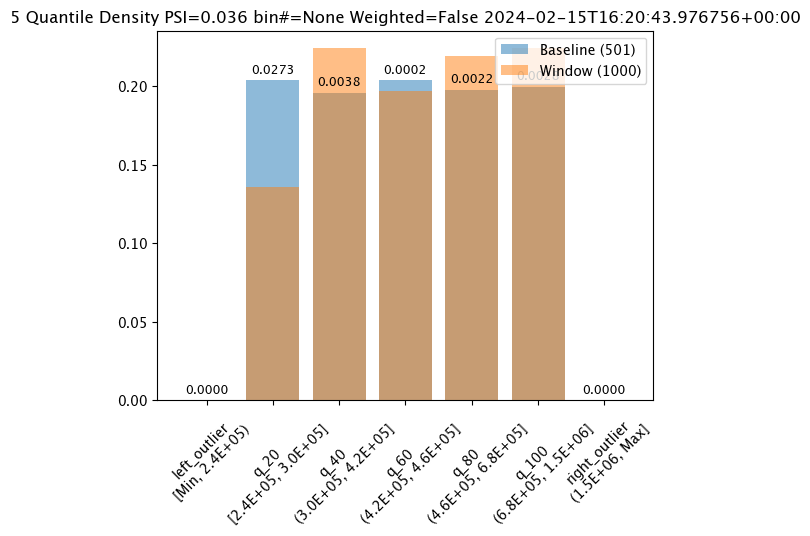
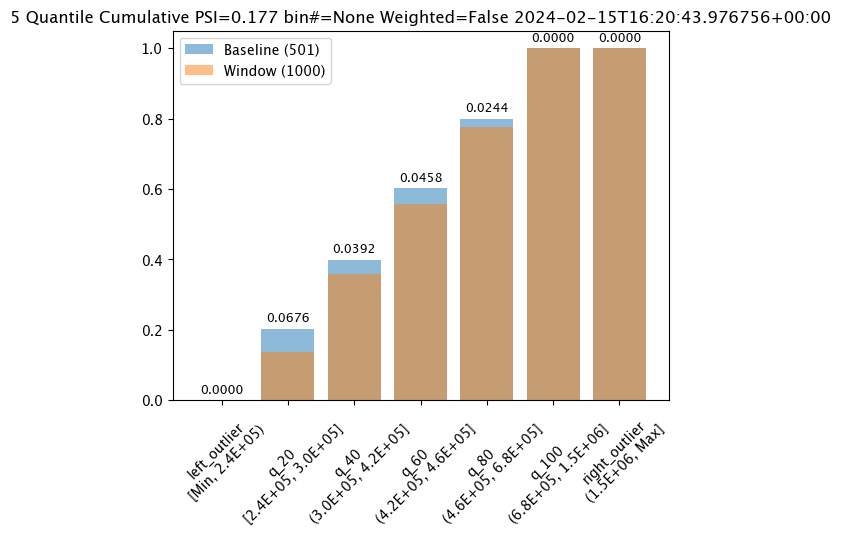
#Aggregation.CUMULATIVE
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
assay_builder_from_dates.summarizer_builder.add_aggregation(wallaroo.assay_config.Aggregation.CUMULATIVE)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Cumulative
metric = PSI
weighted = False
score = 0.17698802395209584
scores = [0.0, 0.06759281437125747, 0.03920159680638724, 0.04579441117764471, 0.02439920159680642, 0.0, 0.0]
index = None
Inference Interval and Inference Width
The inference interval aka window interval sets how often to run the assay analysis. This is set from the wallaroo.assay_config.AssayBuilder.window_builder.add_interval method to collect data expressed in time units: “hours=24”, “minutes=1”, etc.
For example, with an interval of 1 minute, the assay collects data every minute. Within an hour, 60 intervals of data is collected.
We can adjust the interval and see how the assays change based on how frequently they are run.
The width sets the time period from the wallaroo.assay_config.AssayBuilder.window_builder.add_width method to collect data expressed in time units: “hours=24”, “minutes=1”, etc.
For example, an interval of 1 minute and a width of 1 minute collects 1 minutes worth of data every minute. An interval of 1 minute with a width of 5 minutes collects 5 minute of inference data every minute.
By default, the interval and width is 24 hours.
For this example, we’ll adjust the width and interval from 1 minute to 5 minutes and see how the number of analyses and their score changes.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.chart_scores()

# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=5).add_interval(minutes=5).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.chart_scores()
Add Run Until and Add Inference Start
For previewing assays, setting wallaroo.assay_config.AssayBuilder.add_run_until sets the end date and time for collecting inference data. When an assay is uploaded, this setting is no longer valid - assays run at the Inference Interval until the assay is paused.
Setting the wallaroo.assay_config.WindowBuilder.add_start sets the start date and time to collect inference data. When an assay is uploaded, this setting is included, and assay results will be displayed starting from that start date at the Inference Interval until the assay is paused. By default, add_start begins 24 hours after the assay is uploaded unless set in the assay configuration manually.
For the following example, the add_run_until setting is set to datetime.datetime.now() to collect all inference data from assay_window_start up until now, and the second example limits that example to only two minutes of data.
# inference data that includes all of the data until now
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(datetime.datetime.now())
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.chart_scores()

# inference data that includes all of the data until now
assay_builder_from_dates = wl.build_assay(assay_name="assays from date baseline",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and time period
assay_builder_from_dates.add_run_until(assay_window_start+datetime.timedelta(seconds=120))
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
assay_config_from_dates = assay_builder_from_dates.build()
assay_results_from_dates = assay_config_from_dates.interactive_run()
assay_results_from_dates.chart_scores()
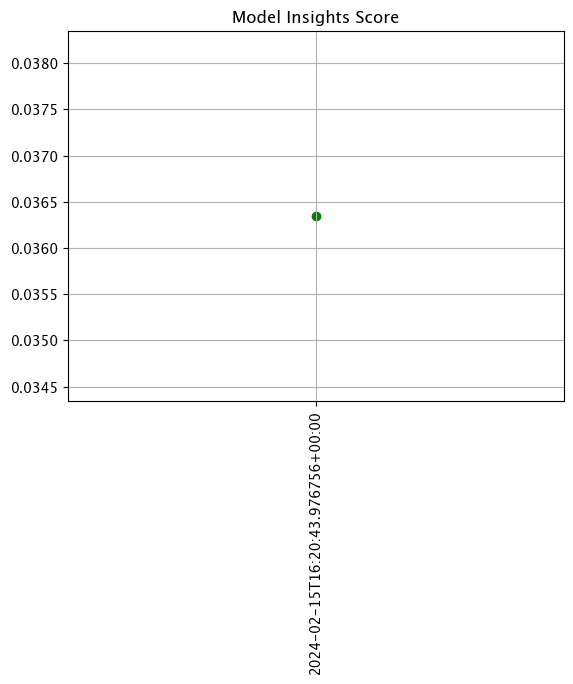
Create Assay
With the assay previewed and configuration options determined, we officially create it by uploading it to the Wallaroo instance.
Once it is uploaded, the assay runs an analysis based on the window width, interval, and the other settings configured.
Assays are uploaded with the wallaroo.assay_config.upload() method. This uploads the assay into the Wallaroo database with the configurations applied and returns the assay id. Note that assay names must be unique across the Wallaroo instance; attempting to upload an assay with the same name as an existing one will return an error.
wallaroo.assay_config.upload() returns the assay id for the assay.
Typically we would just call wallaroo.assay_config.upload() after configuring the assay. For the example below, we will perform the complete configuration in one window to show all of the configuration steps at once before creating the assay.
# Build the assay, based on the start and end of our baseline time,
# and tracking the output variable index 0
assay_builder_from_dates = wl.build_assay(assay_name="assays creation example",
pipeline=mainpipeline,
model_name="house-price-estimator",
iopath="output variable 0",
baseline_start=assay_baseline_start,
baseline_end=assay_baseline_end)
# set the width, interval, and assay start date and time
assay_builder_from_dates.window_builder().add_width(minutes=1).add_interval(minutes=1).add_start(assay_window_start)
# add other options
assay_builder_from_dates.summarizer_builder.add_aggregation(wallaroo.assay_config.Aggregation.CUMULATIVE)
assay_builder_from_dates.summarizer_builder.add_metric(wallaroo.assay_config.Metric.MAXDIFF)
assay_builder_from_dates.add_alert_threshold(0.5)
assay_id = assay_builder_from_dates.upload()
The assay is now visible through the Wallaroo UI by selecting the workspace, then the pipeline, then Insights.
Get Assay Results
Once an assay is created the assay runs an analysis based on the window width, interval, and the other settings configured.
Assay results are retrieved with the wallaroo.client.get_assay_results method, which takes the following parameters:
| Parameter | Type | Description |
|---|---|---|
| assay_id | Integer (Required) | The numerical id of the assay. |
| start | Datetime.Datetime (Required) | The start date and time of historical data from the pipeline to start analyses from. |
| end | Datetime.Datetime (Required) | The end date and time of historical data from the pipeline to limit analyses to. |
- IMPORTANT NOTE: This process requires that additional historical data is generated from the time the assay is created to when the results are available. To add additional inference data, use the Assay Test Data section above.
assay_results = wl.get_assay_results(assay_id=assay_id,
start=assay_window_start,
end=datetime.datetime.now())
assay_results.chart_scores()
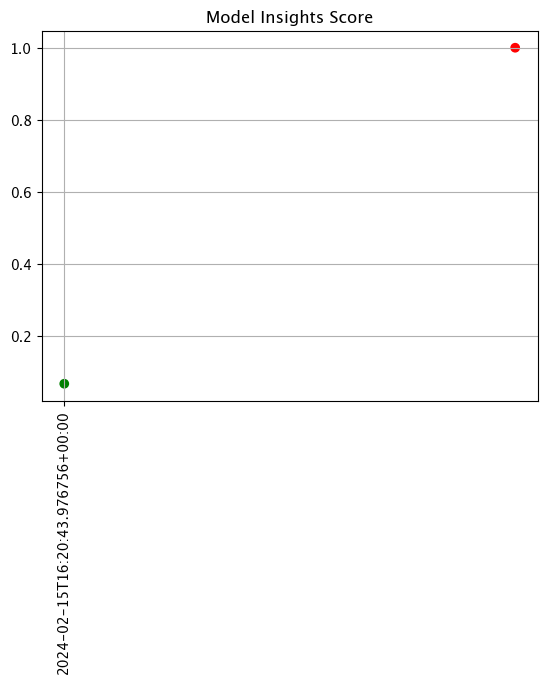
assay_results[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Cumulative
metric = MaxDiff
weighted = False
score = 0.067592815
scores = [0.0, 0.06759281437125747, 0.03920159680638724, 0.04579441117764471, 0.02439920159680642, 0.0, 0.0]
index = 1
List and Retrieve Assay
If the assay id is not already know, it is retrieved from the wallaroo.client.list_assays() method. Select the assay to retrieve data for and retrieve its id with wallaroo.assay.Assay._id method.
wl.list_assays()
| name | active | status | warning_threshold | alert_threshold | pipeline_name |
|---|---|---|---|---|---|
| assays creation example | True | {"run_at": "2024-02-15T16:40:53.212979206+00:00", "num_ok": 0, "num_warnings": 0, "num_alerts": 0} | None | 0.5 | assay-demonstration-tutorial |
retrieved_assay = wl.list_assays()[0]
live_assay_results = wl.get_assay_results(assay_id=retrieved_assay._id,
start=assay_window_start,
end=datetime.datetime.now())
live_assay_results.chart_scores()
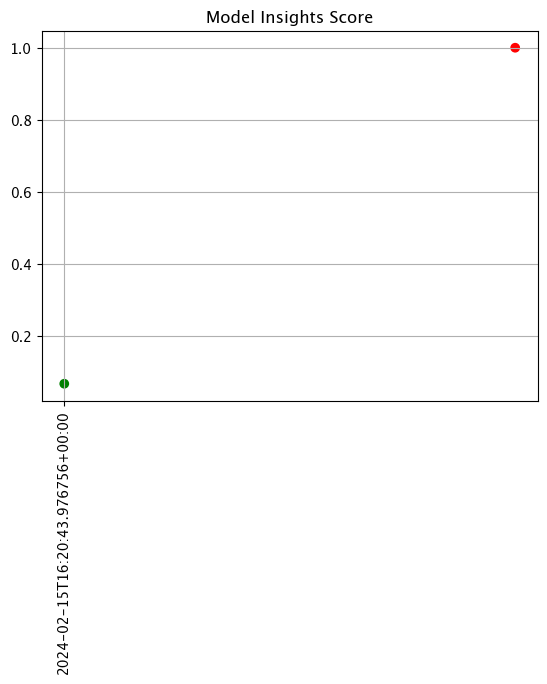
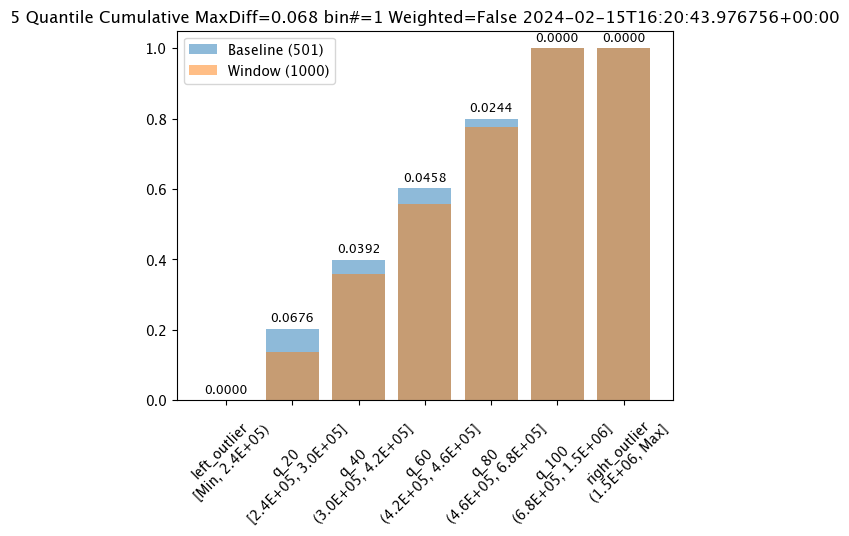
live_assay_results[0].chart()
baseline mean = 495193.23178642715
window mean = 517763.394625
baseline median = 442168.125
window median = 448627.8125
bin_mode = Quantile
aggregation = Cumulative
metric = MaxDiff
weighted = False
score = 0.067592815
scores = [0.0, 0.06759281437125747, 0.03920159680638724, 0.04579441117764471, 0.02439920159680642, 0.0, 0.0]
index = 1
Pause and Resume Assay
Assays are paused and started with the wallaroo.assay.Assay.turn_off and wallaroo.assay.Assay.turn_on methods.
For the following, we retrieve an assay from the wallaroo instance and pause it, then list the assays to verify its setting Active is False.
display(wl.list_assays())
retrieved_assay = wl.list_assays()[0]
| name | active | status | warning_threshold | alert_threshold | pipeline_name |
|---|---|---|---|---|---|
| assays creation example | True | {"run_at": "2024-02-15T16:40:53.212979206+00:00", "num_ok": 0, "num_warnings": 0, "num_alerts": 0} | None | 0.5 | assay-demonstration-tutorial |
Now we pause the assay, and show the assay list to verify it is no longer active.
retrieved_assay.turn_off()
display(wl.list_assays())
| name | active | status | warning_threshold | alert_threshold | pipeline_name |
|---|---|---|---|---|---|
| assays creation example | False | {"run_at": "2024-02-15T16:40:53.212979206+00:00", "num_ok": 0, "num_warnings": 0, "num_alerts": 0} | None | 0.5 | assay-demonstration-tutorial |
We resume the assay and verify its setting Active is True.
retrieved_assay.turn_on()
display(wl.list_assays())
| name | active | status | warning_threshold | alert_threshold | pipeline_name |
|---|---|---|---|---|---|
| assays creation example | True | {"run_at": "2024-02-15T16:40:53.212979206+00:00", "num_ok": 0, "num_warnings": 0, "num_alerts": 0} | None | 0.5 | assay-demonstration-tutorial |