Wallaroo SDK Essentials Guide: Assays Management
Model Insights and Interactive Analysis Introduction
Wallaroo provides the ability to perform interactive analysis so organizations can explore the data from a pipeline and learn how the data is behaving. With this information and the knowledge of your particular business use case you can then choose appropriate thresholds for persistent automatic assays as desired.
IMPORTANT NOTE
Model insights operates over time and is difficult to demo in a notebook without pre-canned data. We assume you have an active pipeline that has been running and making predictions over time and show you the code you may use to analyze your pipeline.
Monitoring tasks called assays monitors a model’s predictions or the data coming into the model against an established baseline. Changes in the distribution of this data can be an indication of model drift, or of a change in the environment that the model trained for. This can provide tips on whether a model needs to be retrained or the environment data analyzed for accuracy or other needs.
Assay Details
Assays contain the following attributes:
| Attribute | Default | Description |
|---|---|---|
| Name | The name of the assay. Assay names must be unique. | |
| Baseline Data | Data that is known to be “typical” (typically distributed) and can be used to determine whether the distribution of new data has changed. | |
| Schedule | Every 24 hours at 1 AM | New assays are configured to run a new analysis for every 24 hours starting at the end of the baseline period. This period can be configured through the SDK. |
| Group Results | Daily | Groups assay results into groups based on either Daily (the default), Weekly, or Monthly. |
| Metric | PSI | Population Stability Index (PSI) is an entropy-based measure of the difference between distributions. Maximum Difference of Bins measures the maximum difference between the baseline and current distributions (as estimated using the bins). Sum of the difference of bins sums up the difference of occurrences in each bin between the baseline and current distributions. |
| Threshold | 0.1 | The threshold for deciding whether the difference between distributions, as evaluated by the above metric, is large (the distributions are different) or small (the distributions are similar). The default of 0.1 is generally a good threshold when using PSI as the metric. |
| Number of Bins | 5 | Sets the number of bins that will be used to partition the baseline data for comparison against how future data falls into these bins. By default, the binning scheme is percentile (quantile) based. The binning scheme can be configured (see Bin Mode, below). Note that the total number of bins will include the set number plus the left_outlier and the right_outlier, so the total number of bins will be the total set + 2. |
| Bin Mode | Quantile | Set the binning scheme. Quantile binning defines the bins using percentile ranges (each bin holds the same percentage of the baseline data). Equal binning defines the bins using equally spaced data value ranges, like a histogram. Custom allows users to set the range of values for each bin, with the Left Outlier always starting at Min (below the minimum values detected from the baseline) and the Right Outlier always ending at Max (above the maximum values detected from the baseline). |
| Bin Weight | Equally Weighted | The bin weights can be either set to Equally Weighted (the default) where each bin is weighted equally, or Custom where the bin weights can be adjusted depending on which are considered more important for detecting model drift. |
Manage Assays via the Wallaroo SDK
List Assays
Assays are listed through the Wallaroo Client list_assays method.
wl.list_assays()
| name | active | status | warning_threshold | alert_threshold | pipeline_name |
|---|---|---|---|---|---|
| api_assay | True | created | 0.0 | 0.1 | housepricepipe |
Interactive Baseline Runs
We can do an interactive run of just the baseline part to see how the baseline data will be put into bins. This assay uses quintiles so all 5 bins (not counting the outlier bins) have 20% of the predictions. We can see the bin boundaries along the x-axis.
baseline_run.chart()
baseline mean = 12.940910643273655
baseline median = 12.884286880493164
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
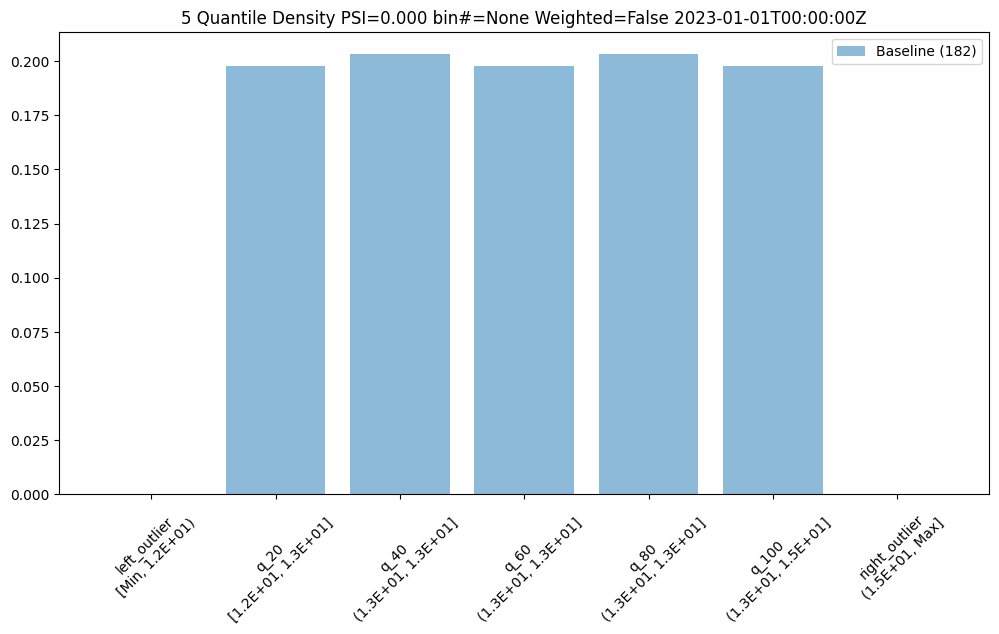
We can also get a dataframe with the bin/edge information.
baseline_run.baseline_bins()
| b_edges | b_edge_names | b_aggregated_values | b_aggregation | |
|---|---|---|---|---|
| 0 | 12.00 | left_outlier | 0.00 | Density |
| 1 | 12.55 | q_20 | 0.20 | Density |
| 2 | 12.81 | q_40 | 0.20 | Density |
| 3 | 12.98 | q_60 | 0.20 | Density |
| 4 | 13.33 | q_80 | 0.20 | Density |
| 5 | 14.97 | q_100 | 0.20 | Density |
| 6 | inf | right_outlier | 0.00 | Density |
The previous assay used quintiles so all of the bins had the same percentage/count of samples. To get bins that are divided equally along the range of values we can use BinMode.EQUAL.
equal_bin_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end)
equal_bin_builder.summarizer_builder.add_bin_mode(BinMode.EQUAL)
equal_baseline = equal_bin_builder.build().interactive_baseline_run()
equal_baseline.chart()
baseline mean = 12.940910643273655
baseline median = 12.884286880493164
bin_mode = Equal
aggregation = Density
metric = PSI
weighted = False
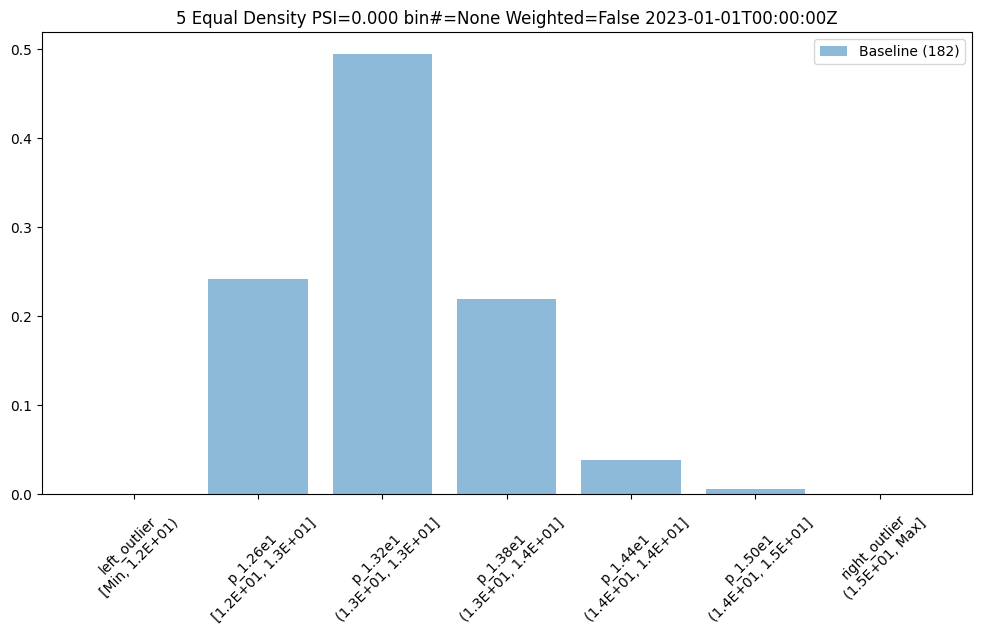
We now see very different bin edges and sample percentages per bin.
equal_baseline.baseline_bins()
| b_edges | b_edge_names | b_aggregated_values | b_aggregation | |
|---|---|---|---|---|
| 0 | 12.00 | left_outlier | 0.00 | Density |
| 1 | 12.60 | p_1.26e1 | 0.24 | Density |
| 2 | 13.19 | p_1.32e1 | 0.49 | Density |
| 3 | 13.78 | p_1.38e1 | 0.22 | Density |
| 4 | 14.38 | p_1.44e1 | 0.04 | Density |
| 5 | 14.97 | p_1.50e1 | 0.01 | Density |
| 6 | inf | right_outlier | 0.00 | Density |
Interactive Assay Runs
By default the assay builder creates an assay with some good starting parameters. In particular the assay is configured to run a new analysis for every 24 hours starting at the end of the baseline period. Additionally, it sets the number of bins to 5 so creates quintiles, and sets the target iopath to "outputs 0 0" which means we want to monitor the first column of the first output/prediction.
We can do an interactive run of just the baseline part to see how the baseline data will be put into bins. This assay uses quintiles so all 5 bins (not counting the outlier bins) have 20% of the predictions. We can see the bin boundaries along the x-axis.
We then run it with interactive_run and convert it to a dataframe for easy analysis with to_dataframe.
Now lets do an interactive run of the first assay as it is configured. Interactive runs don’t save the assay to the database (so they won’t be scheduled in the future) nor do they save the assay results. Instead the results are returned after a short while for further analysis.
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end)
assay_config = assay_builder.add_run_until(last_day).build()
assay_results = assay_config.interactive_run()
assay_df = assay_results.to_dataframe()
assay_df.loc[:, ~assay_df.columns.isin(['assay_id', 'iopath', 'name', 'warning_threshold'])]
| score | start | min | max | mean | median | std | alert_threshold | status | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.00 | 2023-01-02T00:00:00+00:00 | 12.05 | 14.71 | 12.97 | 12.90 | 0.48 | 0.25 | Ok |
| 1 | 0.09 | 2023-01-03T00:00:00+00:00 | 12.04 | 14.65 | 12.96 | 12.93 | 0.41 | 0.25 | Ok |
| 2 | 0.04 | 2023-01-04T00:00:00+00:00 | 11.87 | 14.02 | 12.98 | 12.95 | 0.46 | 0.25 | Ok |
| 3 | 0.06 | 2023-01-05T00:00:00+00:00 | 11.92 | 14.46 | 12.93 | 12.87 | 0.46 | 0.25 | Ok |
| 4 | 0.02 | 2023-01-06T00:00:00+00:00 | 12.02 | 14.15 | 12.95 | 12.90 | 0.43 | 0.25 | Ok |
| 5 | 0.03 | 2023-01-07T00:00:00+00:00 | 12.18 | 14.58 | 12.96 | 12.93 | 0.44 | 0.25 | Ok |
| 6 | 0.02 | 2023-01-08T00:00:00+00:00 | 12.01 | 14.60 | 12.92 | 12.90 | 0.46 | 0.25 | Ok |
| 7 | 0.04 | 2023-01-09T00:00:00+00:00 | 12.01 | 14.40 | 13.00 | 12.97 | 0.45 | 0.25 | Ok |
| 8 | 0.06 | 2023-01-10T00:00:00+00:00 | 11.99 | 14.79 | 12.94 | 12.91 | 0.46 | 0.25 | Ok |
| 9 | 0.02 | 2023-01-11T00:00:00+00:00 | 11.90 | 14.66 | 12.91 | 12.88 | 0.45 | 0.25 | Ok |
| 10 | 0.02 | 2023-01-12T00:00:00+00:00 | 11.96 | 14.82 | 12.94 | 12.90 | 0.46 | 0.25 | Ok |
| 11 | 0.03 | 2023-01-13T00:00:00+00:00 | 12.07 | 14.61 | 12.96 | 12.93 | 0.47 | 0.25 | Ok |
| 12 | 0.15 | 2023-01-14T00:00:00+00:00 | 12.00 | 14.20 | 13.06 | 13.03 | 0.43 | 0.25 | Ok |
| 13 | 2.92 | 2023-01-15T00:00:00+00:00 | 12.74 | 15.62 | 14.00 | 14.01 | 0.57 | 0.25 | Alert |
| 14 | 7.89 | 2023-01-16T00:00:00+00:00 | 14.64 | 17.19 | 15.91 | 15.87 | 0.63 | 0.25 | Alert |
| 15 | 8.87 | 2023-01-17T00:00:00+00:00 | 16.60 | 19.23 | 17.94 | 17.94 | 0.63 | 0.25 | Alert |
| 16 | 8.87 | 2023-01-18T00:00:00+00:00 | 18.67 | 21.29 | 20.01 | 20.04 | 0.64 | 0.25 | Alert |
| 17 | 8.87 | 2023-01-19T00:00:00+00:00 | 20.72 | 23.57 | 22.17 | 22.18 | 0.65 | 0.25 | Alert |
| 18 | 8.87 | 2023-01-20T00:00:00+00:00 | 23.04 | 25.72 | 24.32 | 24.33 | 0.66 | 0.25 | Alert |
| 19 | 8.87 | 2023-01-21T00:00:00+00:00 | 25.06 | 27.67 | 26.48 | 26.49 | 0.63 | 0.25 | Alert |
| 20 | 8.87 | 2023-01-22T00:00:00+00:00 | 27.21 | 29.89 | 28.63 | 28.58 | 0.65 | 0.25 | Alert |
| 21 | 8.87 | 2023-01-23T00:00:00+00:00 | 29.36 | 32.18 | 30.82 | 30.80 | 0.67 | 0.25 | Alert |
| 22 | 8.87 | 2023-01-24T00:00:00+00:00 | 31.56 | 34.35 | 32.98 | 32.98 | 0.65 | 0.25 | Alert |
| 23 | 8.87 | 2023-01-25T00:00:00+00:00 | 33.68 | 36.44 | 35.14 | 35.14 | 0.66 | 0.25 | Alert |
| 24 | 8.87 | 2023-01-26T00:00:00+00:00 | 35.93 | 38.51 | 37.31 | 37.33 | 0.65 | 0.25 | Alert |
| 25 | 3.69 | 2023-01-27T00:00:00+00:00 | 12.06 | 39.91 | 29.29 | 38.65 | 12.66 | 0.25 | Alert |
| 26 | 0.05 | 2023-01-28T00:00:00+00:00 | 11.87 | 13.88 | 12.92 | 12.90 | 0.38 | 0.25 | Ok |
| 27 | 0.10 | 2023-01-29T00:00:00+00:00 | 12.02 | 14.36 | 12.98 | 12.96 | 0.38 | 0.25 | Ok |
| 28 | 0.11 | 2023-01-30T00:00:00+00:00 | 11.99 | 14.44 | 12.89 | 12.88 | 0.37 | 0.25 | Ok |
| 29 | 0.01 | 2023-01-31T00:00:00+00:00 | 12.00 | 14.64 | 12.92 | 12.89 | 0.40 | 0.25 | Ok |
Basic functionality for creating quick charts is included.
assay_results.chart_scores()
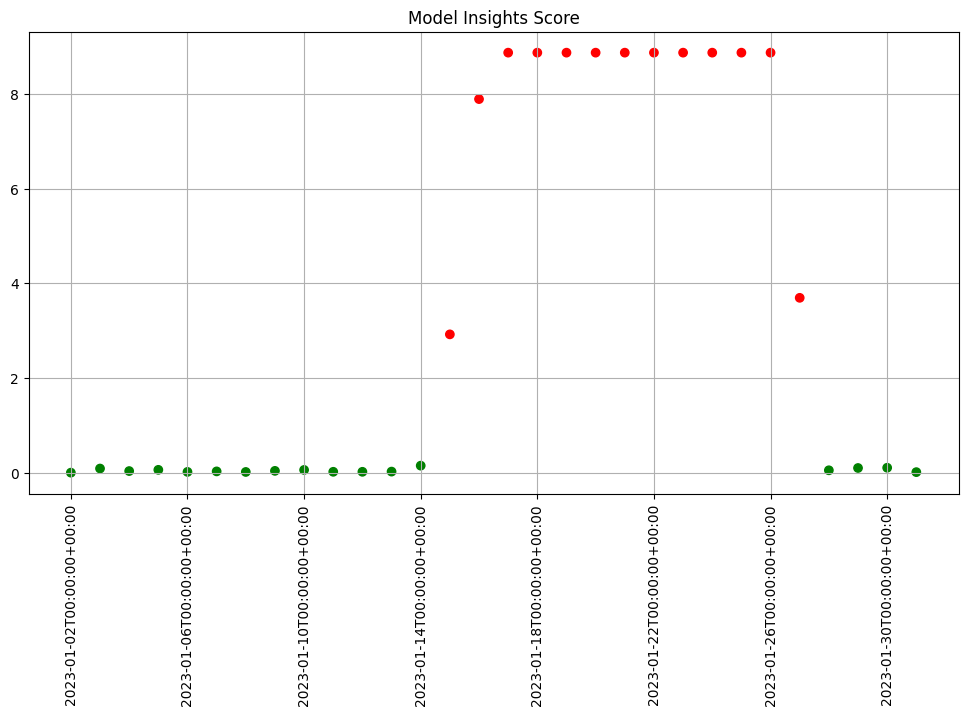
We see that the difference scores are low for a while and then jump up to indicate there is an issue. We can examine that particular window to help us decide if that threshold is set correctly or not.
We can generate a quick chart of the results. This chart shows the 5 quantile bins (quintiles) derived from the baseline data plus one for left outliers and one for right outliers. We also see that the data from the window falls within the baseline quintiles but in a different proportion and is skewing higher. Whether this is an issue or not is specific to your use case.
First lets examine a day that is only slightly different than the baseline. We see that we do see some values that fall outside of the range from the baseline values, the left and right outliers, and that the bin values are different but similar.
assay_results[0].chart()
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0029273068646199748
scores = [0.0, 0.000514261205558409, 0.0002139202456922972, 0.0012617897456473992, 0.0002139202456922972, 0.0007234154220295724, 0.0]
index = None
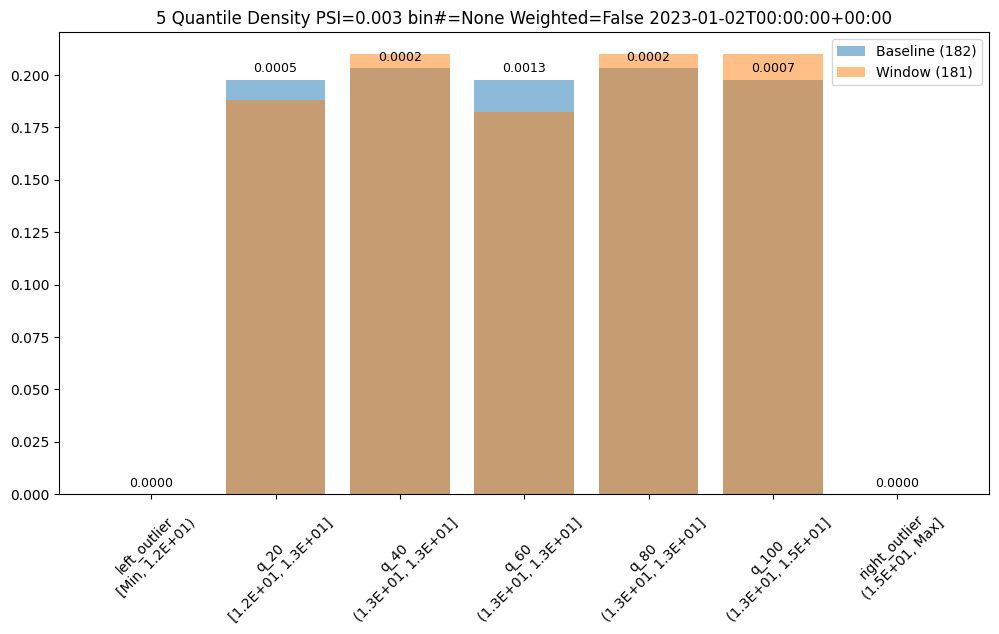
Other days, however are significantly different.
assay_results[12].chart()
baseline mean = 12.940910643273655
window mean = 13.06380216891949
baseline median = 12.884286880493164
window median = 13.027600288391112
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.15060511096978788
scores = [4.6637149189075455e-05, 0.05969428191167242, 0.00806617426854112, 0.008316273402678306, 0.07090885609902021, 0.003572888138686759, 0.0]
index = None
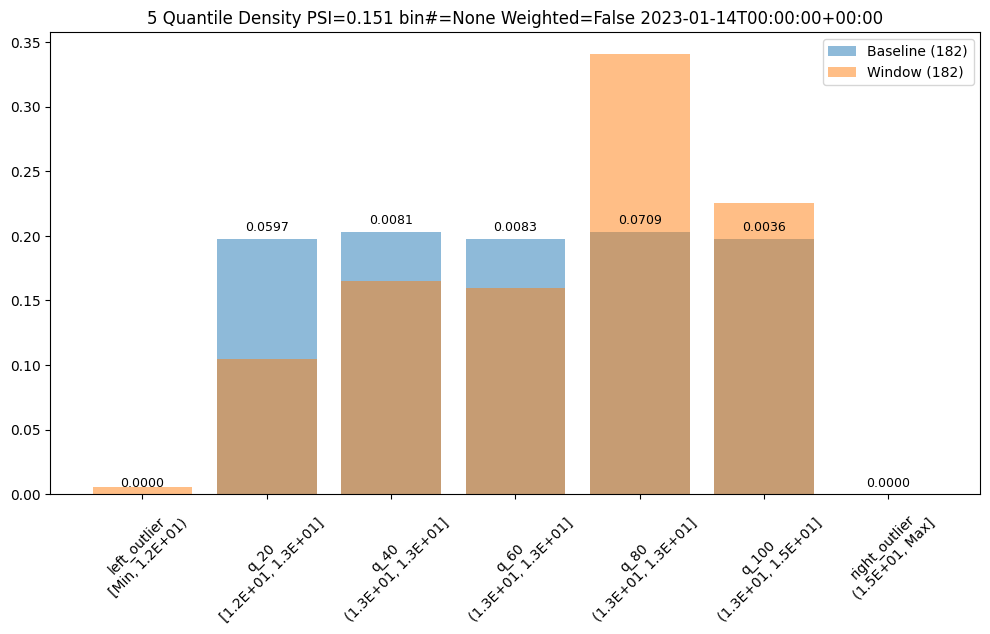
assay_results[13].chart()
baseline mean = 12.940910643273655
window mean = 14.004728427908038
baseline median = 12.884286880493164
window median = 14.009637832641602
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 2.9220486095961196
scores = [0.0, 0.7090936334784107, 0.7130482300184766, 0.33500731896676245, 0.12171058214520876, 0.9038825518183468, 0.1393062931689142]
index = None
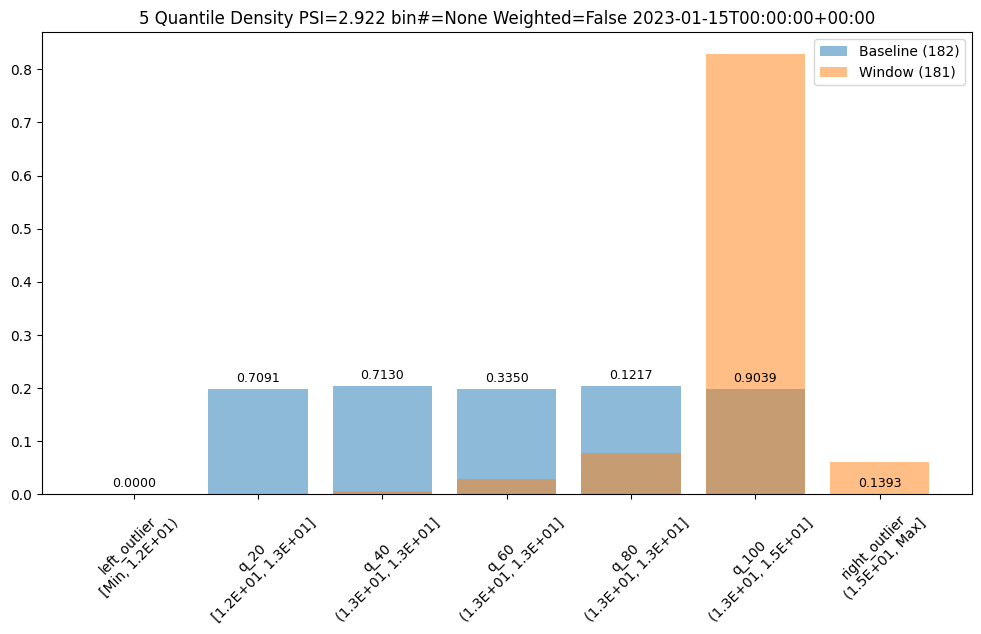
If we want to investigate further, we can run interactive assays on each of the inputs to see if any of them show anything abnormal. In this example we’ll provide the feature labels to create more understandable titles.
The current assay expects continuous data. Sometimes categorical data is encoded as 1 or 0 in a feature and sometimes in a limited number of values such as 1, 2, 3. If one value has high a percentage the analysis emits a warning so that we know the scores for that feature may not behave as we expect.
labels = ['bedrooms', 'bathrooms', 'lat', 'long', 'waterfront', 'sqft_living', 'sqft_lot', 'floors', 'view', 'condition', 'grade', 'sqft_above', 'sqft_basement', 'yr_built', 'yr_renovated', 'sqft_living15', 'sqft_lot15']
topic = wl.get_topic_name(pipeline.id())
all_inferences = wl.get_raw_pipeline_inference_logs(topic, baseline_start, last_day, model_name, limit=1_000_000)
assay_builder = wl.build_assay("Input Assay", pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.window_builder().add_width(hours=4)
assay_config = assay_builder.build()
assay_results = assay_config.interactive_input_run(all_inferences, labels)
iadf = assay_results.to_dataframe()
display(iadf.loc[:, ~iadf.columns.isin(['assay_id', 'iopath', 'name', 'warning_threshold'])])
column distinct_vals label largest_pct
0 17 bedrooms 0.4244
1 44 bathrooms 0.2398
2 3281 lat 0.0014
3 959 long 0.0066
4 4 waterfront 0.9156 *** May not be continuous feature
5 3901 sqft_living 0.0032
6 3487 sqft_lot 0.0173
7 11 floors 0.4567
8 10 view 0.8337
9 9 condition 0.5915
10 19 grade 0.3943
11 745 sqft_above 0.0096
12 309 sqft_basement 0.5582
13 224 yr_built 0.0239
14 77 yr_renovated 0.8889
15 649 sqft_living15 0.0093
16 3280 sqft_lot15 0.0199
| score | start | min | max | mean | median | std | alert_threshold | status | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.19 | 2023-01-02T00:00:00+00:00 | -2.54 | 1.75 | 0.21 | 0.68 | 0.99 | 0.25 | Ok |
| 1 | 0.03 | 2023-01-02T04:00:00+00:00 | -1.47 | 2.82 | 0.21 | -0.40 | 0.95 | 0.25 | Ok |
| 2 | 0.09 | 2023-01-02T08:00:00+00:00 | -2.54 | 3.89 | -0.04 | -0.40 | 1.22 | 0.25 | Ok |
| 3 | 0.05 | 2023-01-02T12:00:00+00:00 | -1.47 | 2.82 | -0.12 | -0.40 | 0.94 | 0.25 | Ok |
| 4 | 0.08 | 2023-01-02T16:00:00+00:00 | -1.47 | 1.75 | -0.00 | -0.40 | 0.76 | 0.25 | Ok |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3055 | 0.08 | 2023-01-31T04:00:00+00:00 | -0.42 | 4.87 | 0.25 | -0.17 | 1.13 | 0.25 | Ok |
| 3056 | 0.58 | 2023-01-31T08:00:00+00:00 | -0.43 | 2.01 | -0.04 | -0.21 | 0.48 | 0.25 | Alert |
| 3057 | 0.13 | 2023-01-31T12:00:00+00:00 | -0.32 | 7.75 | 0.30 | -0.20 | 1.57 | 0.25 | Ok |
| 3058 | 0.26 | 2023-01-31T16:00:00+00:00 | -0.43 | 5.88 | 0.19 | -0.18 | 1.17 | 0.25 | Alert |
| 3059 | 0.84 | 2023-01-31T20:00:00+00:00 | -0.40 | 0.52 | -0.17 | -0.25 | 0.18 | 0.25 | Alert |
3060 rows × 9 columns
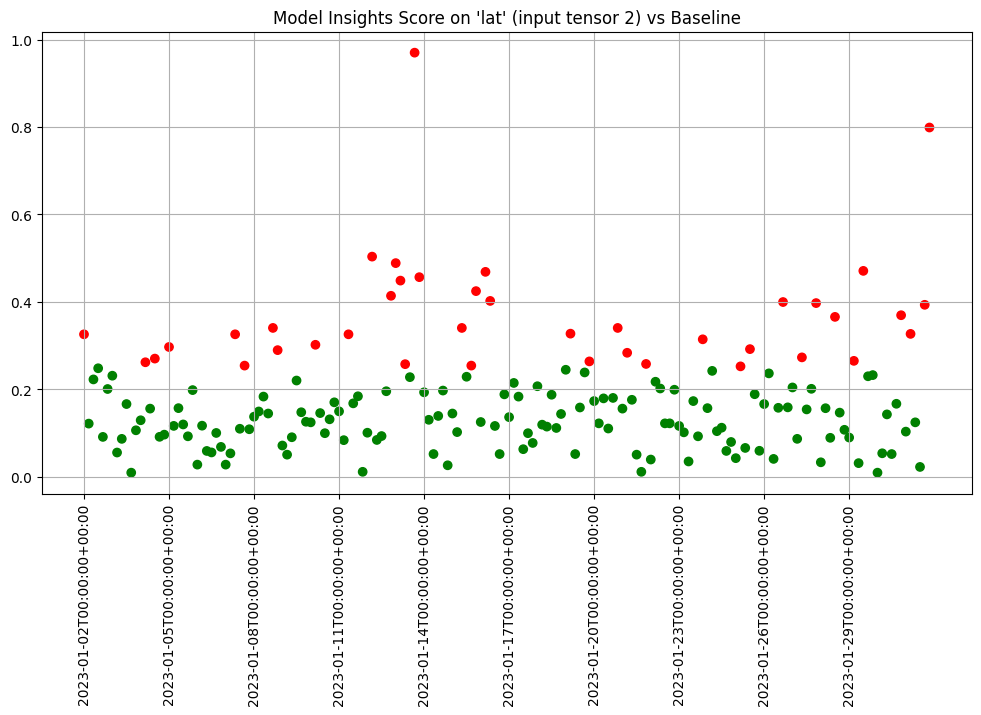
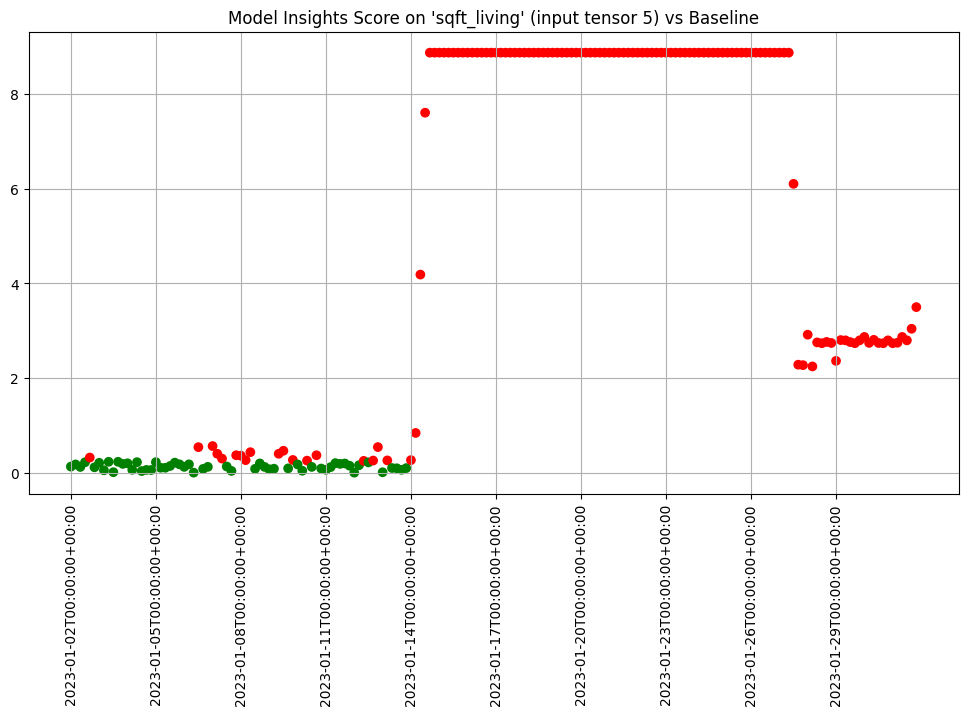
We can chart each of the iopaths and do a visual inspection. From the charts we see that if any of the input features had significant differences in the first two days which we can choose to inspect further. Here we choose to show 3 charts just to save space in this notebook.
assay_results.chart_iopaths(labels=labels, selected_labels=['bedrooms', 'lat', 'sqft_living'])
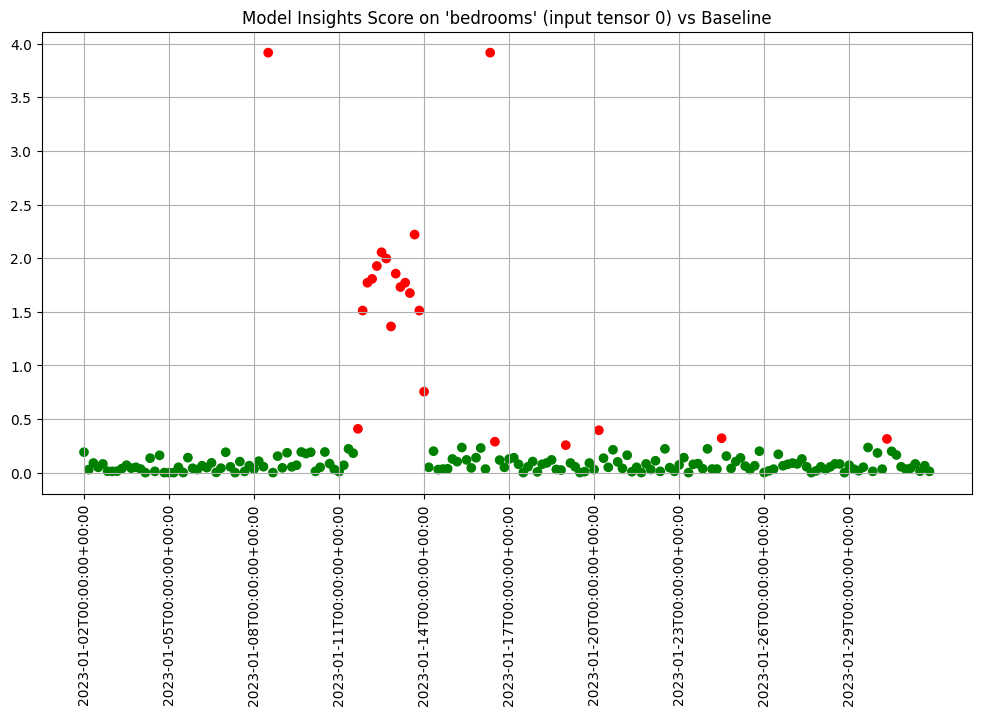
When we are comfortable with what alert threshold should be for our specific purposes we can create and save an assay that will be automatically run on a daily basis.
In this example we’re create an assay that runs everyday against the baseline and has an alert threshold of 0.5.
Once we upload it it will be saved and scheduled for future data as well as run against past data.
alert_threshold = 0.5
import string
import random
prefix= ''.join(random.choice(string.ascii_lowercase) for i in range(4))
assay_name = f"{prefix}example assay"
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_alert_threshold(alert_threshold)
assay_id = assay_builder.upload()
After a short while, we can get the assay results for further analysis.
When we get the assay results, we see that the assays analysis is similar to the interactive run we started with though the analysis for the third day does not exceed the new alert threshold we set. And since we called upload instead of interactive_run the assay was saved to the system and will continue to run automatically on schedule from now on.
Scheduling Assays
By default assays are scheduled to run every 24 hours starting immediately after the baseline period ends.
However, you can control the start time by setting start and the frequency by setting interval on the window.
So to recap:
- The window width is the size of the window. The default is 24 hours.
- The interval is how often the analysis is run, how far the window is slid into the future based on the last run. The default is the window width.
- The window start is when the analysis should start. The default is the end of the baseline period.
For example to run an analysis every 12 hours on the previous 24 hours of data you’d set the window width to 24 (the default) and the interval to 12.
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end)
assay_builder = assay_builder.add_run_until(last_day)
assay_builder.window_builder().add_width(hours=24).add_interval(hours=12)
assay_config = assay_builder.build()
assay_results = assay_config.interactive_run()
print(f"Generated {len(assay_results)} analyses")
Generated 59 analyses
assay_results.chart_scores()
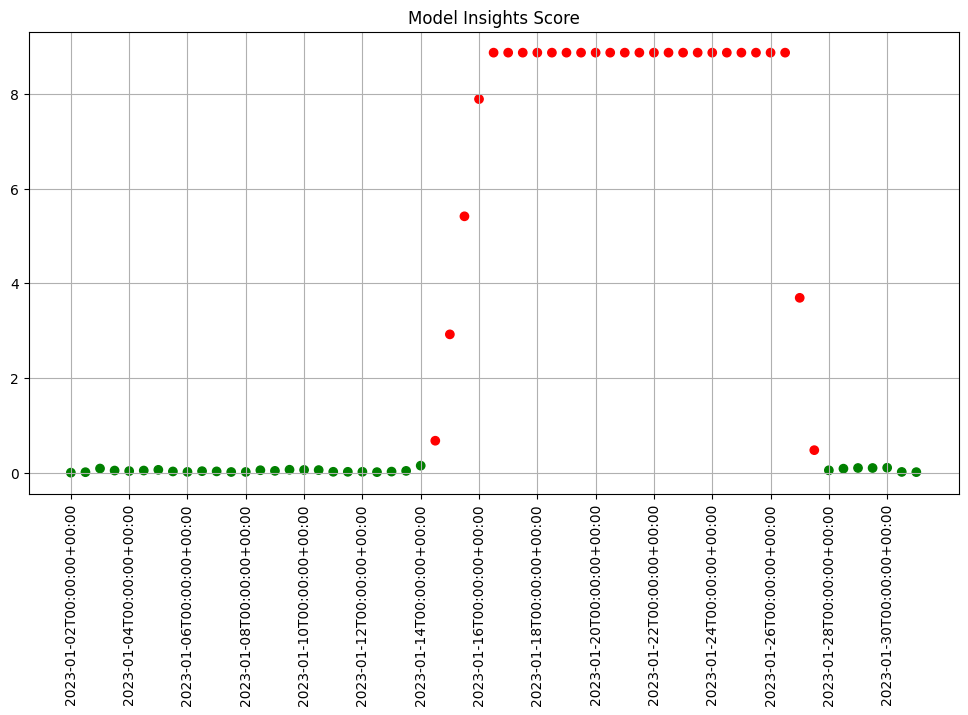
To start a weekly analysis of the previous week on a specific day, set the start date (taking care to specify the desired timezone), and the width and interval to 1 week and of course an analysis won’t be generated till a window is complete.
report_start = datetime.datetime.fromisoformat('2022-01-03T00:00:00+00:00')
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end)
assay_builder = assay_builder.add_run_until(last_day)
assay_builder.window_builder().add_width(weeks=1).add_interval(weeks=1).add_start(report_start)
assay_config = assay_builder.build()
assay_results = assay_config.interactive_run()
print(f"Generated {len(assay_results)} analyses")
Generated 5 analyses
assay_results.chart_scores()
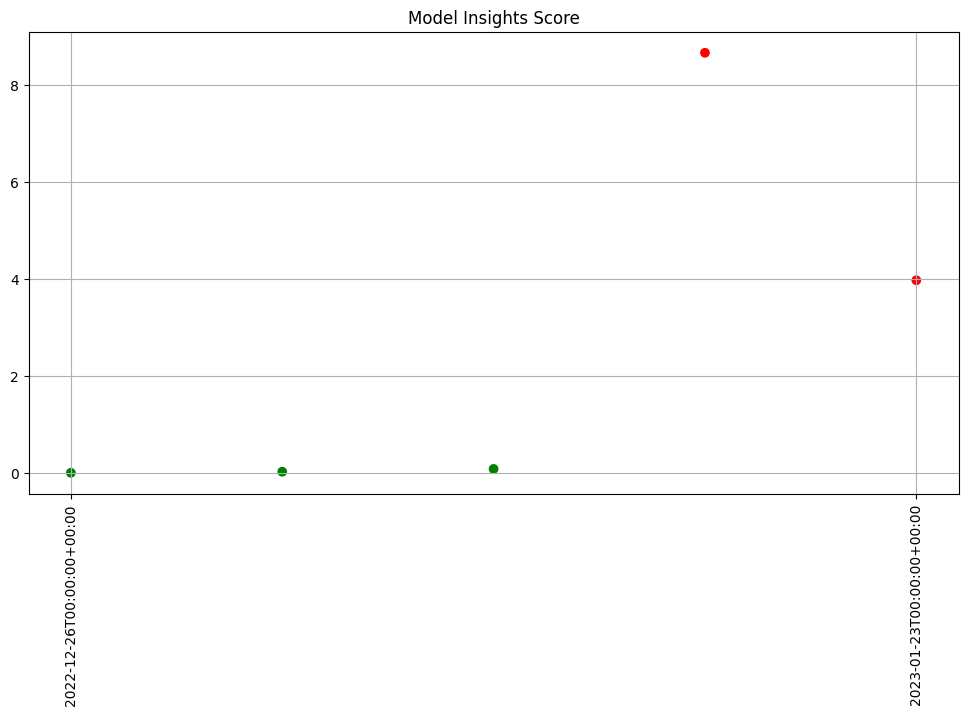
Advanced Configuration
The assay can be configured in a variety of ways to help customize it to your particular needs. Specifically you can:
- change the BinMode to evenly spaced, quantile or user provided
- change the number of bins to use
- provide weights to use when scoring the bins
- calculate the score using the sum of differences, maximum difference or population stability index
- change the value aggregation for the bins to density, cumulative or edges
Lets take a look at these in turn.
Default configuration
First lets look at the default configuration. This is a lot of information but much of it is useful to know where it is available.
We see that the assay is broken up into 4 sections. A top level meta data section, a section for the baseline specification, a section for the window specification and a section that specifies the summarization configuration.
In the meta section we see the name of the assay, that it runs on the first column of the first output "outputs 0 0" and that there is a default threshold of 0.25.
The summarizer section shows us the defaults of Quantile, Density and PSI on 5 bins.
The baseline section shows us that it is configured as a fixed baseline with the specified start and end date times.
And the window tells us what model in the pipeline we are analyzing and how often.
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
print(assay_builder.build().to_json())
{
"name": "onmyexample assay",
"pipeline_id": 1,
"pipeline_name": "housepricepipe",
"active": true,
"status": "created",
"iopath": "output dense_2 0",
"baseline": {
"Fixed": {
"pipeline": "housepricepipe",
"model": "housepricemodel",
"start_at": "2023-01-01T00:00:00+00:00",
"end_at": "2023-01-02T00:00:00+00:00"
}
},
"window": {
"pipeline": "housepricepipe",
"model": "housepricemodel",
"width": "24 hours",
"start": null,
"interval": null
},
"summarizer": {
"type": "UnivariateContinuous",
"bin_mode": "Quantile",
"aggregation": "Density",
"metric": "PSI",
"num_bins": 5,
"bin_weights": null,
"bin_width": null,
"provided_edges": null,
"add_outlier_edges": true
},
"warning_threshold": null,
"alert_threshold": 0.25,
"run_until": "2023-02-01T00:00:00+00:00",
"workspace_id": 5
}
Defaults
We can run the assay interactively and review the first analysis. The method compare_basic_stats gives us a dataframe with basic stats for the baseline and window data.
assay_results = assay_builder.build().interactive_run()
ar = assay_results[0]
ar.compare_basic_stats()
| Baseline | Window | diff | pct_diff | |
|---|---|---|---|---|
| count | 182.00 | 181.00 | -1.00 | -0.55 |
| min | 12.00 | 12.05 | 0.04 | 0.36 |
| max | 14.97 | 14.71 | -0.26 | -1.71 |
| mean | 12.94 | 12.97 | 0.03 | 0.22 |
| median | 12.88 | 12.90 | 0.01 | 0.12 |
| std | 0.45 | 0.48 | 0.03 | 5.68 |
| start | 2023-01-01T00:00:00+00:00 | 2023-01-02T00:00:00+00:00 | NaN | NaN |
| end | 2023-01-02T00:00:00+00:00 | 2023-01-03T00:00:00+00:00 | NaN | NaN |
The method compare_bins gives us a dataframe with the bin information. Such as the number of bins, the right edges, suggested bin/edge names and the values for each bin in the baseline and the window.
assay_bins = ar.compare_bins()
display(assay_bins.loc[:, assay_bins.columns!='w_aggregation'])
| b_edges | b_edge_names | b_aggregated_values | b_aggregation | w_edges | w_edge_names | w_aggregated_values | diff_in_pcts | |
|---|---|---|---|---|---|---|---|---|
| 0 | 12.00 | left_outlier | 0.00 | Density | 12.00 | left_outlier | 0.00 | 0.00 |
| 1 | 12.55 | q_20 | 0.20 | Density | 12.55 | e_1.26e1 | 0.19 | -0.01 |
| 2 | 12.81 | q_40 | 0.20 | Density | 12.81 | e_1.28e1 | 0.21 | 0.01 |
| 3 | 12.98 | q_60 | 0.20 | Density | 12.98 | e_1.30e1 | 0.18 | -0.02 |
| 4 | 13.33 | q_80 | 0.20 | Density | 13.33 | e_1.33e1 | 0.21 | 0.01 |
| 5 | 14.97 | q_100 | 0.20 | Density | 14.97 | e_1.50e1 | 0.21 | 0.01 |
| 6 | NaN | right_outlier | 0.00 | Density | NaN | right_outlier | 0.00 | 0.00 |
We can also plot the chart to visualize the values of the bins.
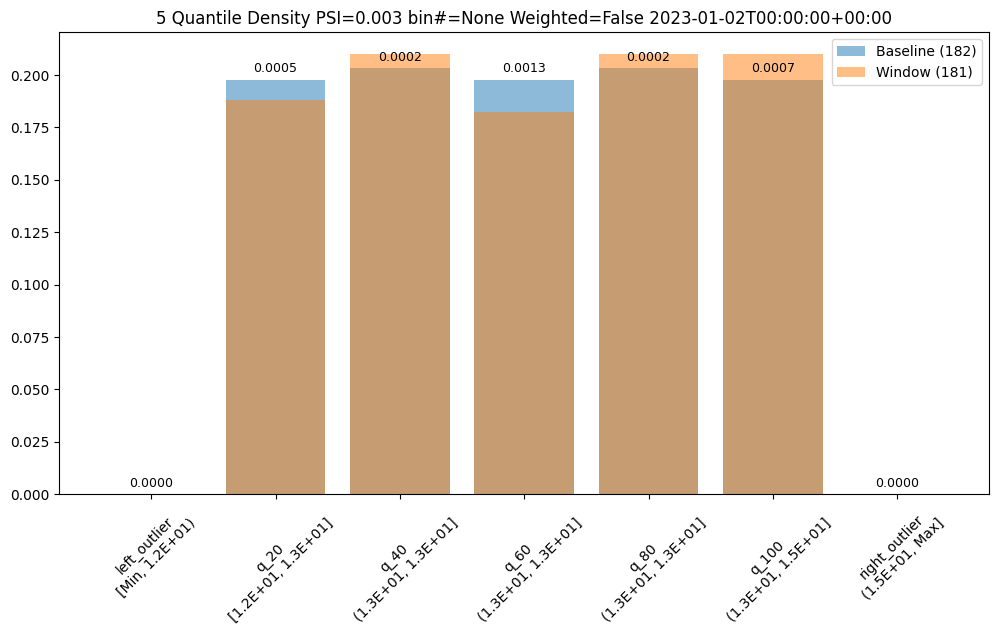
ar.chart()
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0029273068646199748
scores = [0.0, 0.000514261205558409, 0.0002139202456922972, 0.0012617897456473992, 0.0002139202456922972, 0.0007234154220295724, 0.0]
index = None
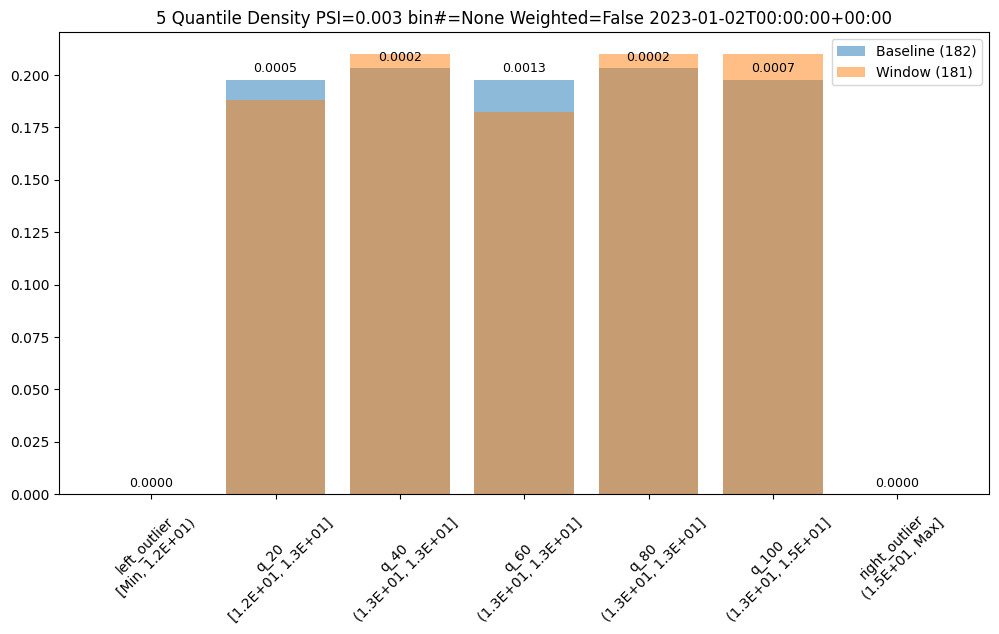
Binning Mode
We can change the bin mode algorithm to equal and see that the bins/edges are partitioned at different points and the bins have different values.
prefix= ''.join(random.choice(string.ascii_lowercase) for i in range(4))
assay_name = f"{prefix}example assay"
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.summarizer_builder.add_bin_mode(BinMode.EQUAL)
assay_results = assay_builder.build().interactive_run()
assay_results_df = assay_results[0].compare_bins()
display(assay_results_df.loc[:, ~assay_results_df.columns.isin(['b_aggregation', 'w_aggregation'])])
assay_results[0].chart()
| b_edges | b_edge_names | b_aggregated_values | w_edges | w_edge_names | w_aggregated_values | diff_in_pcts | |
|---|---|---|---|---|---|---|---|
| 0 | 12.00 | left_outlier | 0.00 | 12.00 | left_outlier | 0.00 | 0.00 |
| 1 | 12.60 | p_1.26e1 | 0.24 | 12.60 | e_1.26e1 | 0.24 | 0.00 |
| 2 | 13.19 | p_1.32e1 | 0.49 | 13.19 | e_1.32e1 | 0.48 | -0.02 |
| 3 | 13.78 | p_1.38e1 | 0.22 | 13.78 | e_1.38e1 | 0.22 | -0.00 |
| 4 | 14.38 | p_1.44e1 | 0.04 | 14.38 | e_1.44e1 | 0.06 | 0.02 |
| 5 | 14.97 | p_1.50e1 | 0.01 | 14.97 | e_1.50e1 | 0.01 | 0.00 |
| 6 | NaN | right_outlier | 0.00 | NaN | right_outlier | 0.00 | 0.00 |
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Equal
aggregation = Density
metric = PSI
weighted = False
score = 0.011074287819376092
scores = [0.0, 7.3591419975306595e-06, 0.000773779195360713, 8.538514991838585e-05, 0.010207597078872246, 1.6725322721660374e-07, 0.0]
index = None
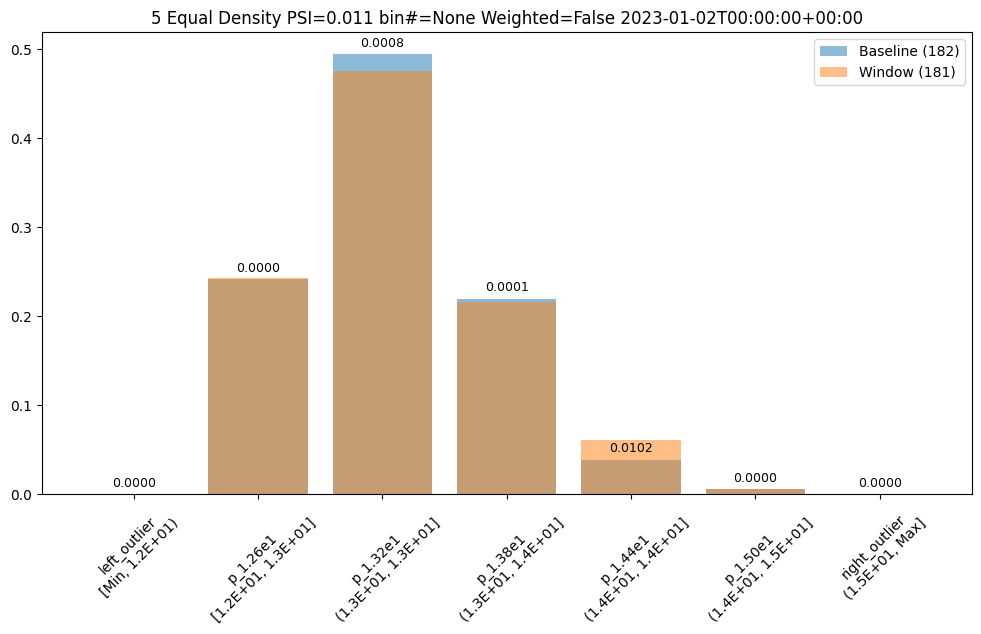
User Provided Bin Edges
The values in this dataset run from ~11.6 to ~15.81. And lets say we had a business reason to use specific bin edges. We can specify them with the BinMode.PROVIDED and specifying a list of floats with the right hand / upper edge of each bin and optionally the lower edge of the smallest bin. If the lowest edge is not specified the threshold for left outliers is taken from the smallest value in the baseline dataset.
edges = [11.0, 12.0, 13.0, 14.0, 15.0, 16.0]
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.summarizer_builder.add_bin_mode(BinMode.PROVIDED, edges)
assay_results = assay_builder.build().interactive_run()
assay_results_df = assay_results[0].compare_bins()
display(assay_results_df.loc[:, ~assay_results_df.columns.isin(['b_aggregation', 'w_aggregation'])])
assay_results[0].chart()
| b_edges | b_edge_names | b_aggregated_values | w_edges | w_edge_names | w_aggregated_values | diff_in_pcts | |
|---|---|---|---|---|---|---|---|
| 0 | 11.00 | left_outlier | 0.00 | 11.00 | left_outlier | 0.00 | 0.00 |
| 1 | 12.00 | e_1.20e1 | 0.00 | 12.00 | e_1.20e1 | 0.00 | 0.00 |
| 2 | 13.00 | e_1.30e1 | 0.62 | 13.00 | e_1.30e1 | 0.59 | -0.03 |
| 3 | 14.00 | e_1.40e1 | 0.36 | 14.00 | e_1.40e1 | 0.35 | -0.00 |
| 4 | 15.00 | e_1.50e1 | 0.02 | 15.00 | e_1.50e1 | 0.06 | 0.03 |
| 5 | 16.00 | e_1.60e1 | 0.00 | 16.00 | e_1.60e1 | 0.00 | 0.00 |
| 6 | NaN | right_outlier | 0.00 | NaN | right_outlier | 0.00 | 0.00 |
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Provided
aggregation = Density
metric = PSI
weighted = False
score = 0.0321620386600679
scores = [0.0, 0.0, 0.0014576920813015586, 3.549754401142936e-05, 0.030668849034754912, 0.0, 0.0]
index = None
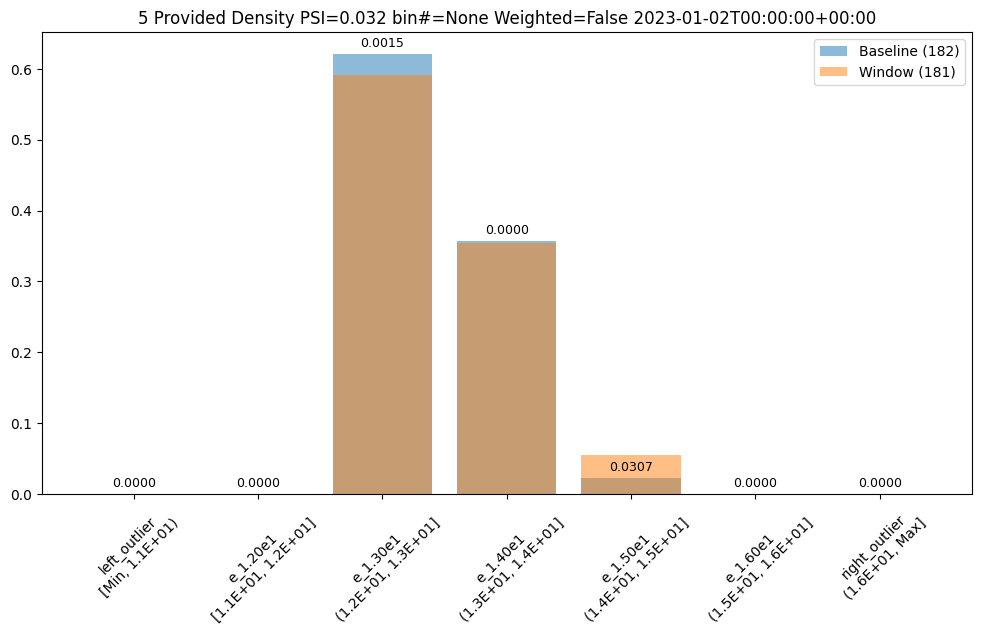
Number of Bins
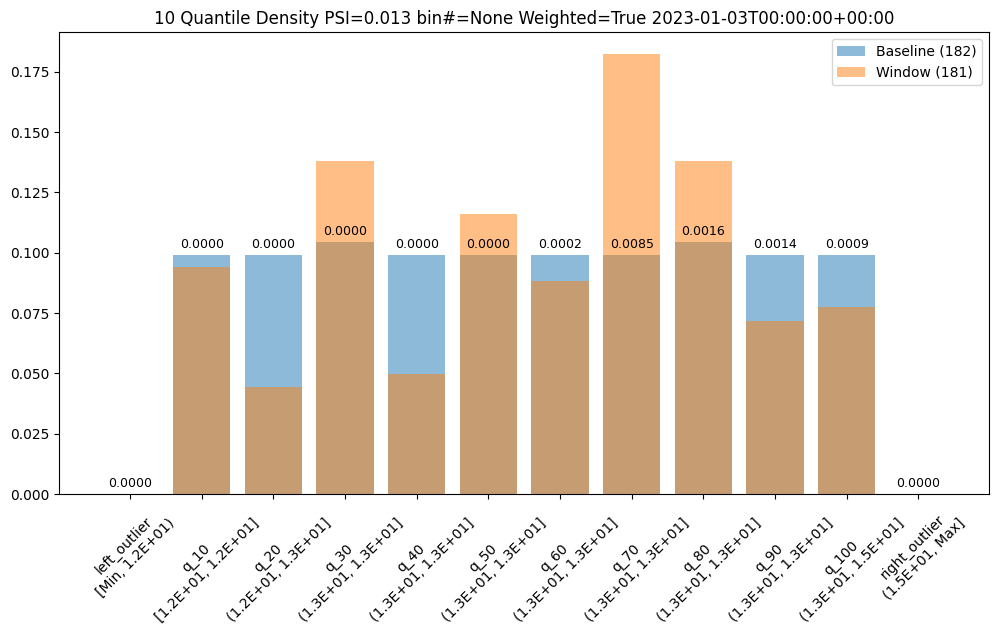
We could also choose to a different number of bins, lets say 10, which can be evenly spaced or based on the quantiles (deciles).
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.summarizer_builder.add_bin_mode(BinMode.QUANTILE).add_num_bins(10)
assay_results = assay_builder.build().interactive_run()
assay_results_df = assay_results[1].compare_bins()
display(assay_results_df.loc[:, ~assay_results_df.columns.isin(['b_aggregation', 'w_aggregation'])])
assay_results[1].chart()
| b_edges | b_edge_names | b_aggregated_values | w_edges | w_edge_names | w_aggregated_values | diff_in_pcts | |
|---|---|---|---|---|---|---|---|
| 0 | 12.00 | left_outlier | 0.00 | 12.00 | left_outlier | 0.00 | 0.00 |
| 1 | 12.41 | q_10 | 0.10 | 12.41 | e_1.24e1 | 0.09 | -0.00 |
| 2 | 12.55 | q_20 | 0.10 | 12.55 | e_1.26e1 | 0.04 | -0.05 |
| 3 | 12.72 | q_30 | 0.10 | 12.72 | e_1.27e1 | 0.14 | 0.03 |
| 4 | 12.81 | q_40 | 0.10 | 12.81 | e_1.28e1 | 0.05 | -0.05 |
| 5 | 12.88 | q_50 | 0.10 | 12.88 | e_1.29e1 | 0.12 | 0.02 |
| 6 | 12.98 | q_60 | 0.10 | 12.98 | e_1.30e1 | 0.09 | -0.01 |
| 7 | 13.15 | q_70 | 0.10 | 13.15 | e_1.32e1 | 0.18 | 0.08 |
| 8 | 13.33 | q_80 | 0.10 | 13.33 | e_1.33e1 | 0.14 | 0.03 |
| 9 | 13.47 | q_90 | 0.10 | 13.47 | e_1.35e1 | 0.07 | -0.03 |
| 10 | 14.97 | q_100 | 0.10 | 14.97 | e_1.50e1 | 0.08 | -0.02 |
| 11 | NaN | right_outlier | 0.00 | NaN | right_outlier | 0.00 | 0.00 |
baseline mean = 12.940910643273655
window mean = 12.956829186961135
baseline median = 12.884286880493164
window median = 12.929338455200195
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.16591076620684958
scores = [0.0, 0.0002571306027792045, 0.044058279699182114, 0.009441459631493015, 0.03381618572319047, 0.0027335446937028877, 0.0011792419836838435, 0.051023062424253904, 0.009441459631493015, 0.008662563542113508, 0.0052978382749576496, 0.0]
index = None
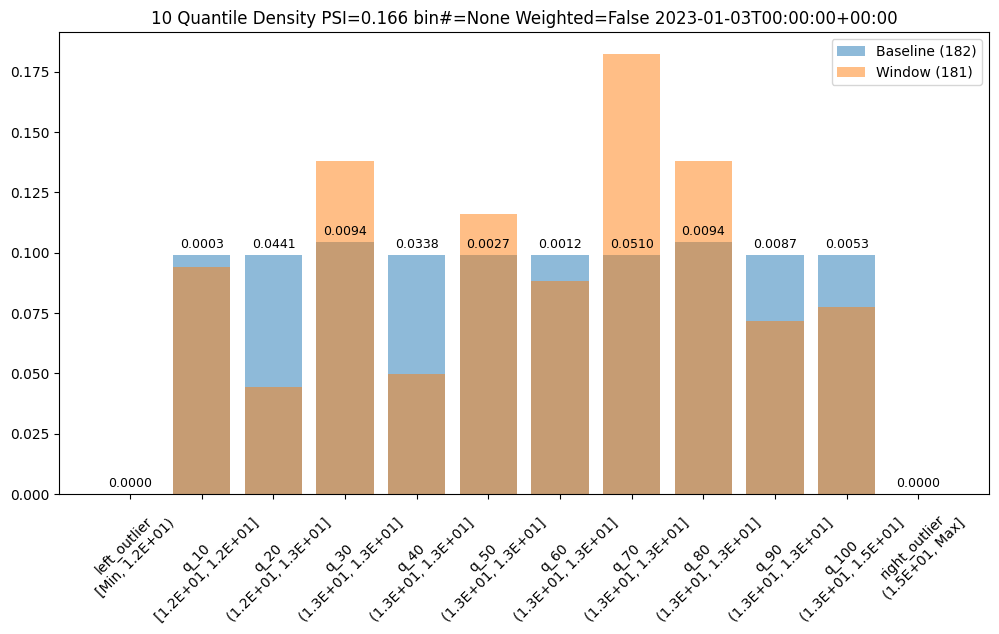
Bin Weights
Now lets say we only care about differences at the higher end of the range. We can use weights to specify that difference in the lower bins should not be counted in the score.
If we stick with 10 bins we can provide 10 a vector of 12 weights. One weight each for the original bins plus one at the front for the left outlier bin and one at the end for the right outlier bin.
Note we still show the values for the bins but the scores for the lower 5 and left outlier are 0 and only the right half is counted and reflected in the score.
weights = [0] * 6
weights.extend([1] * 6)
print("Using weights: ", weights)
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.summarizer_builder.add_bin_mode(BinMode.QUANTILE).add_num_bins(10).add_bin_weights(weights)
assay_results = assay_builder.build().interactive_run()
assay_results_df = assay_results[1].compare_bins()
display(assay_results_df.loc[:, ~assay_results_df.columns.isin(['b_aggregation', 'w_aggregation'])])
assay_results[1].chart()
Using weights: [0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1]
| b_edges | b_edge_names | b_aggregated_values | w_edges | w_edge_names | w_aggregated_values | diff_in_pcts | |
|---|---|---|---|---|---|---|---|
| 0 | 12.00 | left_outlier | 0.00 | 12.00 | left_outlier | 0.00 | 0.00 |
| 1 | 12.41 | q_10 | 0.10 | 12.41 | e_1.24e1 | 0.09 | -0.00 |
| 2 | 12.55 | q_20 | 0.10 | 12.55 | e_1.26e1 | 0.04 | -0.05 |
| 3 | 12.72 | q_30 | 0.10 | 12.72 | e_1.27e1 | 0.14 | 0.03 |
| 4 | 12.81 | q_40 | 0.10 | 12.81 | e_1.28e1 | 0.05 | -0.05 |
| 5 | 12.88 | q_50 | 0.10 | 12.88 | e_1.29e1 | 0.12 | 0.02 |
| 6 | 12.98 | q_60 | 0.10 | 12.98 | e_1.30e1 | 0.09 | -0.01 |
| 7 | 13.15 | q_70 | 0.10 | 13.15 | e_1.32e1 | 0.18 | 0.08 |
| 8 | 13.33 | q_80 | 0.10 | 13.33 | e_1.33e1 | 0.14 | 0.03 |
| 9 | 13.47 | q_90 | 0.10 | 13.47 | e_1.35e1 | 0.07 | -0.03 |
| 10 | 14.97 | q_100 | 0.10 | 14.97 | e_1.50e1 | 0.08 | -0.02 |
| 11 | NaN | right_outlier | 0.00 | NaN | right_outlier | 0.00 | 0.00 |
baseline mean = 12.940910643273655
window mean = 12.956829186961135
baseline median = 12.884286880493164
window median = 12.929338455200195
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = True
score = 0.012600694309416988
scores = [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.00019654033061397393, 0.00850384373737565, 0.0015735766052488358, 0.0014437605903522511, 0.000882973045826275, 0.0]
index = None
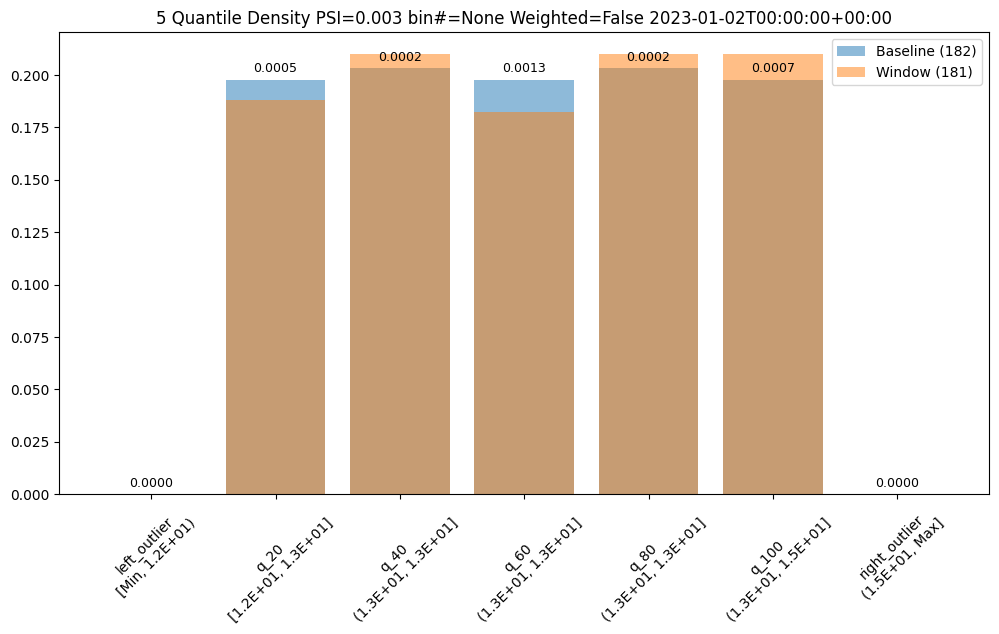
Metrics
The score is a distance or dis-similarity measure. The larger it is the less similar the two distributions are. We currently support
summing the differences of each individual bin, taking the maximum difference and a modified Population Stability Index (PSI).
The following three charts use each of the metrics. Note how the scores change. The best one will depend on your particular use case.
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_results = assay_builder.build().interactive_run()
assay_results[0].chart()
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0029273068646199748
scores = [0.0, 0.000514261205558409, 0.0002139202456922972, 0.0012617897456473992, 0.0002139202456922972, 0.0007234154220295724, 0.0]
index = None
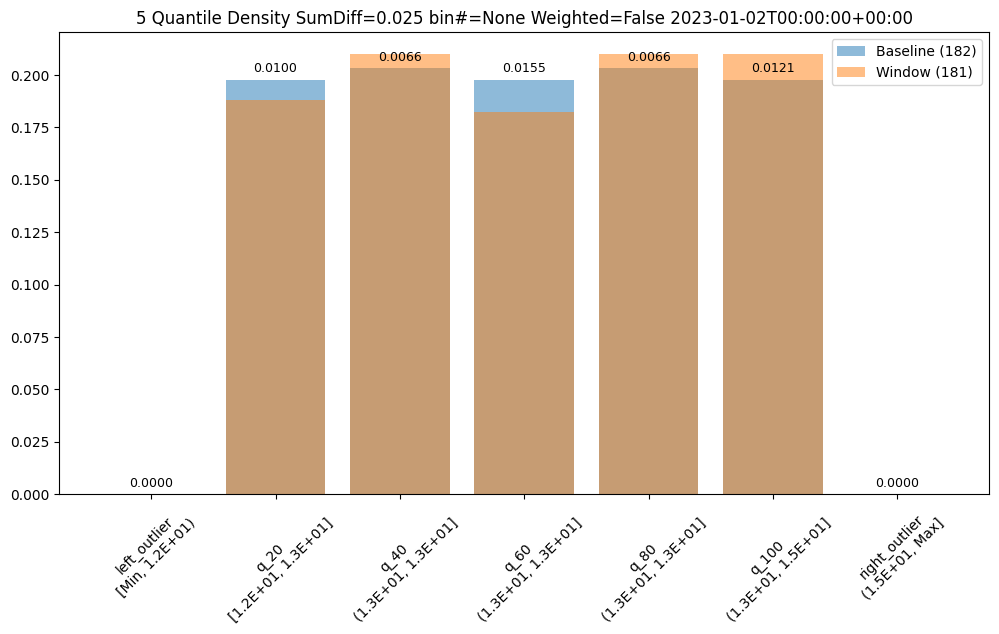
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.summarizer_builder.add_metric(Metric.SUMDIFF)
assay_results = assay_builder.build().interactive_run()
assay_results[0].chart()
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Quantile
aggregation = Density
metric = SumDiff
weighted = False
score = 0.025438649748041997
scores = [0.0, 0.009956893934794486, 0.006648048084512165, 0.01548175581324751, 0.006648048084512165, 0.012142553579017668, 0.0]
index = None
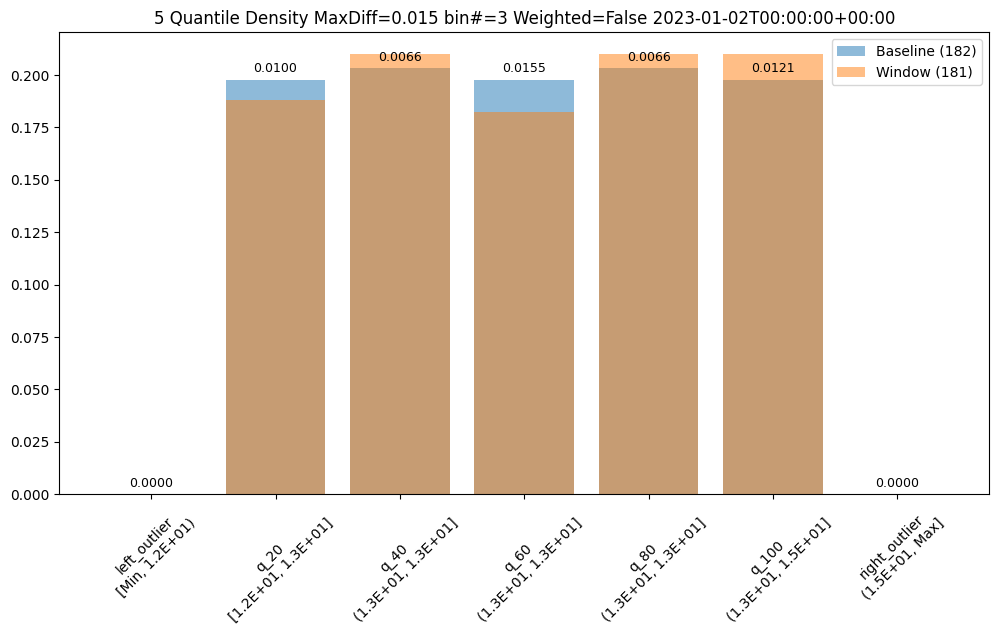
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.summarizer_builder.add_metric(Metric.MAXDIFF)
assay_results = assay_builder.build().interactive_run()
assay_results[0].chart()
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Quantile
aggregation = Density
metric = MaxDiff
weighted = False
score = 0.01548175581324751
scores = [0.0, 0.009956893934794486, 0.006648048084512165, 0.01548175581324751, 0.006648048084512165, 0.012142553579017668, 0.0]
index = 3
Aggregation Options
Also, bin aggregation can be done in histogram Aggregation.DENSITY style (the default) where we count the number/percentage of values that fall in each bin or Empirical Cumulative Density Function style Aggregation.CUMULATIVE where we keep a cumulative count of the values/percentages that fall in each bin.
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
assay_builder.summarizer_builder.add_aggregation(Aggregation.DENSITY)
assay_results = assay_builder.build().interactive_run()
assay_results[0].chart()
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Quantile
aggregation = Density
metric = PSI
weighted = False
score = 0.0029273068646199748
scores = [0.0, 0.000514261205558409, 0.0002139202456922972, 0.0012617897456473992, 0.0002139202456922972, 0.0007234154220295724, 0.0]
index = None
assay_builder = wl.build_assay(assay_name, pipeline, model_name, baseline_start, baseline_end).add_run_until(last_day)
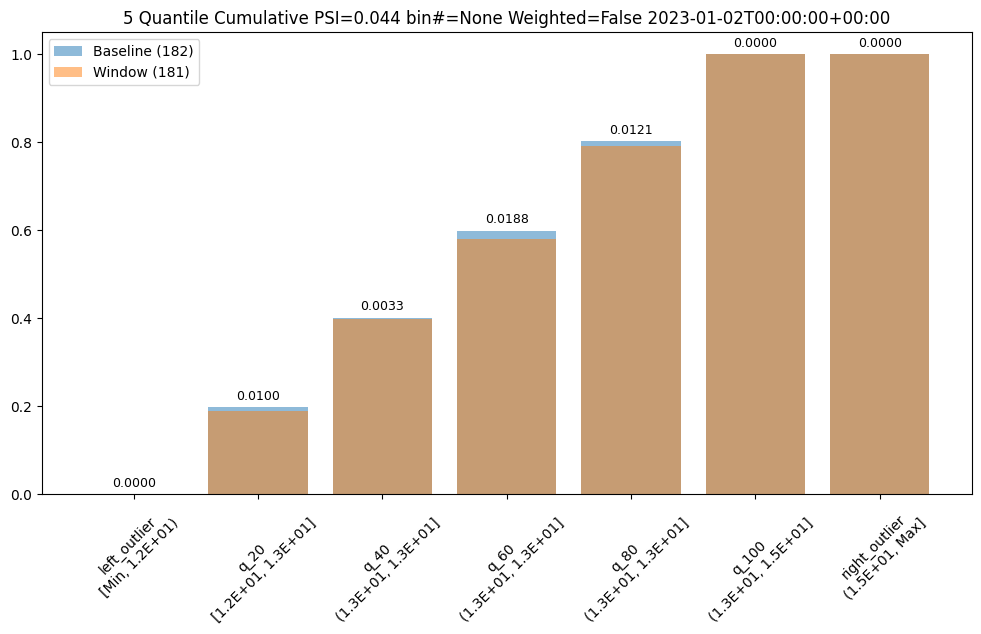
assay_builder.summarizer_builder.add_aggregation(Aggregation.CUMULATIVE)
assay_results = assay_builder.build().interactive_run()
assay_results[0].chart()
baseline mean = 12.940910643273655
window mean = 12.969964654406132
baseline median = 12.884286880493164
window median = 12.899214744567873
bin_mode = Quantile
aggregation = Cumulative
metric = PSI
weighted = False
score = 0.04419889502762442
scores = [0.0, 0.009956893934794486, 0.0033088458502823492, 0.01879060166352986, 0.012142553579017725, 0.0, 0.0]
index = None